2MTZ
 
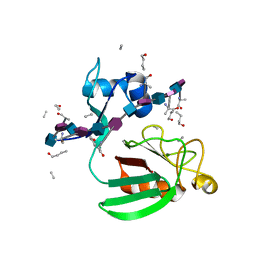 | | Haddock model of Bacillus subtilis L,D-transpeptidase in complex with a peptidoglycan hexamuropeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, Putative L,D-transpeptidase YkuD, intact bacterial peptidoglycan | | Authors: | Schanda, P, Triboulet, S, Laguri, C, Bougault, C, Ayala, I, Callon, M, Arthur, M, Simorre, J. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic model of a cell-wall cross-linking enzyme in complex with an intact bacterial peptidoglycan.
J.Am.Chem.Soc., 136, 2014
|
|
9HG8
 
 | | BAM-SurA complex in the swing-out state with BamC resolved | | Descriptor: | Chaperone SurA, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Lehner, P.A, Jakob, R.P, Hiller, S. | | Deposit date: | 2024-11-19 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture and conformational dynamics of the BAM-SurA holo insertase complex.
Sci Adv, 11, 2025
|
|
9HG6
 
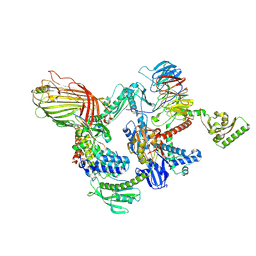 | | BAM-SurA complex in the swing-in state | | Descriptor: | Chaperone SurA, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Lehner, P.A, Jakob, R.P, Hiller, S. | | Deposit date: | 2024-11-19 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Architecture and conformational dynamics of the BAM-SurA holo insertase complex.
Sci Adv, 11, 2025
|
|
9HG9
 
 | | BAM-SurA-darobactin complex in the swing-in state | | Descriptor: | Chaperone SurA, Darobactin A, MAGNESIUM ION, ... | | Authors: | Lehner, P.A, Jakob, R.P, Hiller, S. | | Deposit date: | 2024-11-19 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Architecture and conformational dynamics of the BAM-SurA holo insertase complex.
Sci Adv, 11, 2025
|
|
9HG7
 
 | | BAM-SurA complex in the swing-out state | | Descriptor: | Chaperone SurA, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Lehner, P.A, Jakob, R.P, Hiller, S. | | Deposit date: | 2024-11-19 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Architecture and conformational dynamics of the BAM-SurA holo insertase complex.
Sci Adv, 11, 2025
|
|
9HG5
 
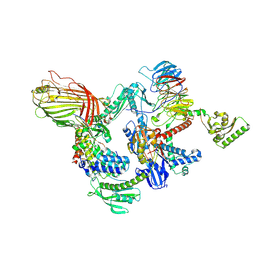 | | BAM-SurA complex in the swing-in state with full length BamC resolved | | Descriptor: | Chaperone SurA, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Lehner, P.A, Jakob, R.P, Hiller, S. | | Deposit date: | 2024-11-19 | | Release date: | 2025-04-16 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Architecture and conformational dynamics of the BAM-SurA holo insertase complex.
Sci Adv, 11, 2025
|
|
9HGA
 
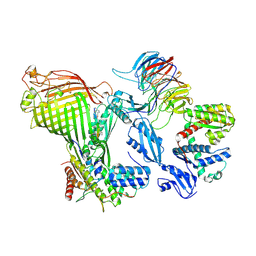 | | BAM-SurA-darobactin complex in the swing-out state | | Descriptor: | Chaperone SurA, Darobactin A, Outer membrane protein assembly factor BamA, ... | | Authors: | Lehner, P.A, Jakob, R.P, Hiller, S. | | Deposit date: | 2024-11-19 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Architecture and conformational dynamics of the BAM-SurA holo insertase complex.
Sci Adv, 11, 2025
|
|
9FLG
 
 | | Solution NMR structure of the Delta60 domain of the hepatitis delta virus small antigen SHDAg | | Descriptor: | Large delta antigen | | Authors: | Yang, Y, Delcourte, L, Fogeron, M.L, Bockmann, A, Lecoq, L. | | Deposit date: | 2024-06-05 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-14 | | Method: | SOLUTION NMR | | Cite: | Structure and nucleic acid interactions of the S Delta 60 domain of the hepatitis delta virus small antigen.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
