2M0N
 
 | |
2LXF
 
 | |
2MYY
 
 | |
2N6G
 
 | |
2MU0
 
 | |
5UJ5
 
 | |
5JHI
 
 | | Solution structure of the de novo mini protein gEHE_06 | | Descriptor: | W35 | | Authors: | Buchko, G.W, Bahl, C.D, Gilmore, J.M, Pulavarti, S.V, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-21 | | Release date: | 2016-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
5JI4
 
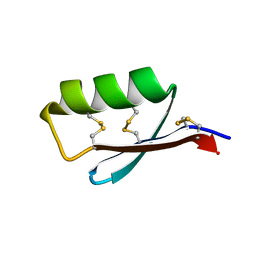 | | Solution structure of the de novo mini protein gEEHE_02 | | Descriptor: | W37 | | Authors: | Buchko, G.W, Bahl, C.D, Pulavarti, S.V, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-21 | | Release date: | 2016-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
2F3L
 
 | |
2G0Y
 
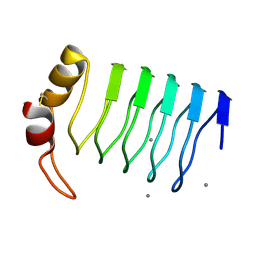 | | Crystal Structure of a Lumenal Pentapeptide Repeat Protein from Cyanothece sp 51142 at 2.3 Angstrom Resolution. Tetragonal Crystal Form | | Descriptor: | CALCIUM ION, pentapeptide repeat protein | | Authors: | Kennedy, M.A, Ni, S, Buchko, G.W, Robinson, H. | | Deposit date: | 2006-02-13 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of two potentially universal turn motifs that shape the repeated five-residues fold - Crystal structure of a lumenal pentapeptide repeat protein from Cyanothece 51142
Protein Sci., 15, 2006
|
|
2LR3
 
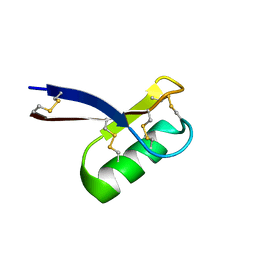 | |
2O5F
 
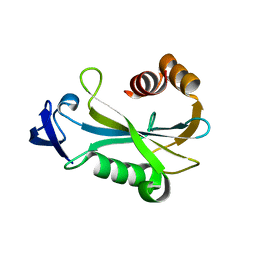 | | Crystal Structure of DR0079 from Deinococcus radiodurans at 1.9 Angstrom Resolution | | Descriptor: | Putative Nudix hydrolase DR_0079 | | Authors: | Kennedy, M.A, Buchko, G.W, Ni, S, Robinson, H. | | Deposit date: | 2006-12-05 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and Structural Characterization of DR_0079 from Deinococcus radiodurans, a Novel Nudix Hydrolase with a Preference for Cytosine (Deoxy)ribonucleoside 5'-Di- and Triphosphates.
Biochemistry, 47, 2008
|
|
2O6W
 
 | | Crystal Structure of a Pentapeptide Repeat Protein (Rfr23) from the cyanobacterium Cyanothece 51142 | | Descriptor: | ARSENIC, Repeat Five Residue (Rfr) protein or pentapeptide repeat protein | | Authors: | Kennedy, M.A, Buchko, G.W, Ni, S, Robinson, H, Pakrasi, H.B. | | Deposit date: | 2006-12-08 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the structural variation between pentapeptide repeat proteins-Crystal structure of Rfr23 from Cyanothece 51142.
J.Struct.Biol., 162, 2008
|
|
3CSX
 
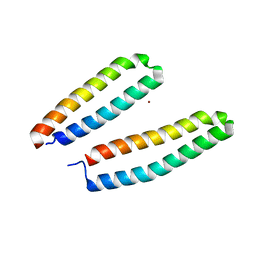 | |
4E98
 
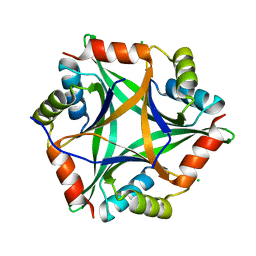 | |
3GMT
 
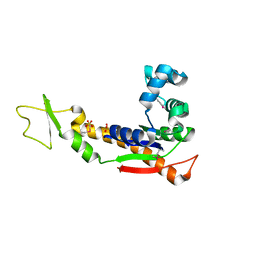 | | Crystal structure of adenylate kinase from burkholderia pseudomallei | | Descriptor: | Adenylate kinase, SULFATE ION | | Authors: | Abendroth, J, Staker, B.L, Robinson, H, Buchko, G.W, Hewitt, S.N, Napuli, A.J, Van Voorhis, W, Stacy, R, Myler, P.J, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-03-15 | | Release date: | 2009-06-02 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of Burkholderia pseudomallei adenylate kinase (Adk): profound asymmetry in the crystal structure of the 'open' state.
Biochem.Biophys.Res.Commun., 394, 2010
|
|
4H6P
 
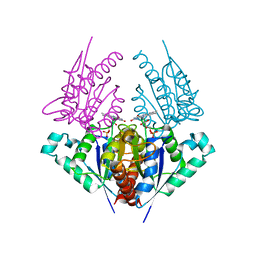 | | Crystal structure of a putative chromate reductase from Gluconacetobacter hansenii, Gh-ChrR, containing a R101A substitution. | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Zhang, Y, Robinson, H, Buchko, G.W. | | Deposit date: | 2012-09-19 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.556 Å) | | Cite: | Mechanistic insights of chromate and uranyl reduction by the NADPH-dependent FMN reductase, ChrR, from Gluconacetobacter hansenii
To be Published
|
|
4HS4
 
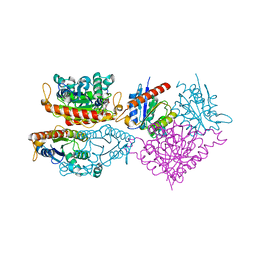 | | Crystal structure of a putative chromate reductase from Gluconacetobacter hansenii, Gh-ChrR, containing a Y129N substitution. | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Zhang, Y, Robinson, H, Buchko, G.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic insights of chromate and uranyl reduction by the NADPH-dependent FMN reductase, ChrR, from Gluconacetobacter hansenii
To be Published
|
|
3NJ2
 
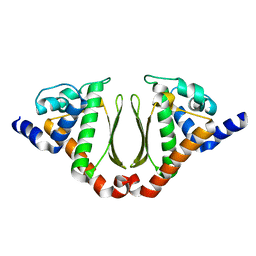 | | Crystal structure of cce_0566 from the cyanobacterium Cyanothece 51142, a protein associated with nitrogen fixation from the DUF269 family | | Descriptor: | DUF269-containing protein | | Authors: | Robinson, H, Ralston, C.Y, Addlagatta, A, Buchko, G.W. | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structure of cce_0566 from Cyanothece 51142, a protein associated with nitrogen fixation in the DUF269 family.
Febs Lett., 586, 2012
|
|
1ALF
 
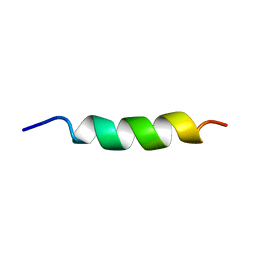 | |
1ALE
 
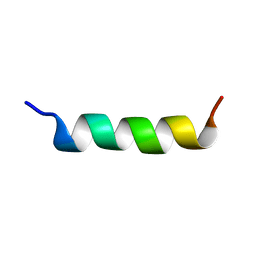 | |
1OPP
 
 | | PEPTIDE OF HUMAN APOLIPOPROTEIN C-I RESIDUES 1-38, NMR, 28 STRUCTURES | | Descriptor: | APOLIPOPROTEIN C-I | | Authors: | Rozek, A, Buchko, G.W, Kanda, P, Cushley, R.J. | | Deposit date: | 1997-05-08 | | Release date: | 1998-05-13 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | Conformational studies of the N-terminal lipid-associating domain of human apolipoprotein C-I by CD and 1H NMR spectroscopy.
Protein Sci., 6, 1997
|
|
1SSV
 
 | | Compensating bends in a 16 base-pair DNA oligomer containing a T3A3 segment | | Descriptor: | 5'-D(*CP*GP*AP*GP*GP*TP*TP*TP*AP*AP*AP*CP*CP*TP*CP*G)-3' | | Authors: | McAteer, K, Aceves-Gaona, A, Michalczyk, R, Buchko, G.W, Isern, N.G, Silks, L.A, Miller, J.H, Kennedy, M.A. | | Deposit date: | 2004-03-24 | | Release date: | 2004-12-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Compensating bends in a 16-base-pair DNA oligomer containing a T(3)A(3) segment: A NMR study of global DNA curvature
Biopolymers, 75, 2004
|
|
1SS7
 
 | | Compensating bends in a 16 base-pair DNA oligomer containing a T3A3 segment | | Descriptor: | 5'-D(*CP*GP*AP*GP*GP*TP*TP*TP*AP*AP*AP*CP*CP*TP*CP*G)-3' | | Authors: | McAteer, K, Aceves-Gaona, A, Michalczyk, R, Buchko, G.W, Isern, N.G, Silks, L.A, Miller, J.H, Kennedy, M.A. | | Deposit date: | 2004-03-23 | | Release date: | 2004-12-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Compensating bends in a 16-base-pair DNA oligomer containing a T(3)A(3) segment: A NMR study of global DNA curvature
Biopolymers, 75, 2004
|
|
6U6G
 
 | |
