1ODG
 
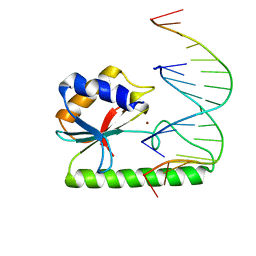 | | Very-short-patch DNA repair endonuclease bound to its reaction product site | | Descriptor: | 5'-D(*TP*AP*GP*GP*CP*5CM*TP*GP*GP*AP*TP*CP)-3', DNA MISMATCH ENDONUCLEASE, ZINC ION | | Authors: | Bunting, K.A, Roe, S.M, Headley, A, Brown, T, Savva, R, Pearl, L.H. | | Deposit date: | 2003-02-19 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Escherichia Coli Dcm Very-Short-Patch DNA Repair Endonuclease Bound to its Reaction Product-Site in a DNA Superhelix
Nucleic Acids Res., 31, 2003
|
|
404D
 
 | |
1DRR
 
 | | DNA/RNA HYBRID DUPLEX CONTAINING A PURINE-RICH DNA STRAND, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*AP*GP*AP*AP*GP*C)-3'), RNA (5'-R(*GP*CP*UP*UP*CP*UP*CP*UP*UP*C)-3') | | Authors: | Gyi, J.I, Lane, A.N, Conn, G.L, Brown, T. | | Deposit date: | 1997-10-21 | | Release date: | 1998-04-22 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes.
Biochemistry, 37, 1998
|
|
1D99
 
 | |
2DBE
 
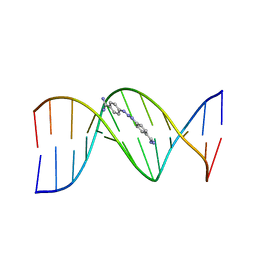 | | CRYSTAL STRUCTURE OF A BERENIL-DODECANUCLEOTIDE COMPLEX: THE ROLE OF WATER IN SEQUENCE-SPECIFIC LIGAND BINDING | | Descriptor: | BERENIL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Brown, D.G, Sanderson, M.R, Skelly, J.V, Jenkins, T.C, Brown, T, Garman, E, Stuart, D.I, Neidle, S. | | Deposit date: | 1990-03-19 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a berenil-dodecanucleotide complex: the role of water in sequence-specific ligand binding.
EMBO J., 9, 1990
|
|
1D92
 
 | | REFINED CRYSTAL STRUCTURE OF AN OCTANUCLEOTIDE DUPLEX WITH G.T MISMATCHED BASE-PAIRS | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*CP*TP*CP*C)-3') | | Authors: | Hunter, W.N, Kneale, G, Brown, T, Rabinovich, D, Kennard, O. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Refined crystal structure of an octanucleotide duplex with G . T mismatched base-pairs.
J.Mol.Biol., 190, 1986
|
|
1RRD
 
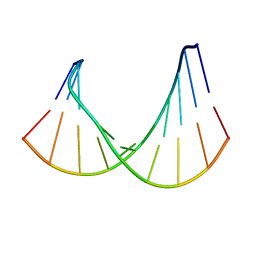 | | DNA/RNA HYBRID DUPLEX CONTAINING A PURINE-RICH RNA STRAND, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*CP*TP*CP*TP*TP*C)-3'), RNA (5'-R(*GP*AP*AP*GP*AP*GP*AP*AP*GP*C)-3') | | Authors: | Gyi, J.I, Lane, A.N, Conn, G.L, Brown, T. | | Deposit date: | 1997-10-21 | | Release date: | 1998-04-22 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes.
Biochemistry, 37, 1998
|
|
1RRR
 
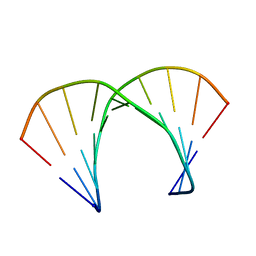 | | RNA DUPLEX CONTAINING A PURINE-RICH STRAND, NMR, 10 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*AP*AP*GP*AP*GP*AP*AP*GP*C)-3'), RNA (5'-R(*GP*CP*UP*UP*CP*UP*CP*UP*UP*C)-3') | | Authors: | Gyi, J.I, Lane, A.N, Conn, G.L, Brown, T. | | Deposit date: | 1997-10-21 | | Release date: | 1998-04-22 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes.
Biochemistry, 37, 1998
|
|
5MVT
 
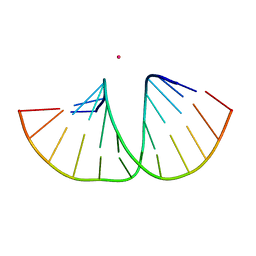 | | Crystal structure of an A-DNA dodecamer featuring an alternating pyrimidine-purine sequence | | Descriptor: | COBALT (III) ION, DNA | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVL
 
 | | Crystal structure of an A-DNA dodecamer containing 5-bromouracil | | Descriptor: | Brominated DNA dodecamer, MAGNESIUM ION | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-16 | | Release date: | 2017-05-10 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.405 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVQ
 
 | | Crystal structure of an unmodified, self-complementary dodecamer. | | Descriptor: | DNA, MAGNESIUM ION | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVU
 
 | |
5MVP
 
 | | Crystal structure of an A-DNA dodecamer containing the GGGCCC motif | | Descriptor: | DNA, POTASSIUM ION | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVK
 
 | |
6QJR
 
 | | Crystal structure of a DNA dodecamer containing a tetramethylpyrrolinoxyl (nitroxide) spin label | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*TP*TP*TP*(5NO)P*GP*CP*G)-3') | | Authors: | Hardwick, J.S, Haugland, M.M, Ptchelkine, D, Brown, T, Anderson, E.E. | | Deposit date: | 2019-01-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 2'-Alkynyl spin-labelling is a minimally perturbing tool for DNA structural analysis.
Nucleic Acids Res., 48, 2020
|
|
1DA9
 
 | | ANTHRACYCLINE-DNA INTERACTIONS AT UNFAVOURABLE BASE BASE-PAIR TRIPLET-BINDING SITES: STRUCTURES OF D(CGGCCG)/DAUNOMYCIN AND D(TGGCCA)/ADRIAMYCIN COMPL | | Descriptor: | DNA (5'-D(*TP*GP*GP*CP*CP*A)-3'), DOXORUBICIN | | Authors: | Leonard, G.A, Hambley, T.W, McAuley-Hecht, K, Brown, T, Hunter, W.N. | | Deposit date: | 1993-01-21 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anthracycline-DNA interactions at unfavourable base-pair triplet-binding sites: structures of d(CGGCCG)/daunomycin and d(TGGCCA)/adriamycin complexes.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1DN6
 
 | | THE CRYSTAL STRUCTURE OF D(GGATGGGAG). AN ESSENTIAL PART OF THE BINDING SITE FOR TRANSCRIPTION FACTOR IIIA | | Descriptor: | DNA (5'-D(*CP*TP*CP*CP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*GP*GP*GP*AP*G)-3') | | Authors: | McCall, M, Brown, T, Hunter, W.N, Kennard, O. | | Deposit date: | 1987-05-11 | | Release date: | 1988-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of d(GGATGGGAG): an essential part of the binding site for transcription factor IIIA.
Nature, 322, 1986
|
|
1D75
 
 | | CONFORMATION OF THE GUANINE.8-OXOADENINE BASE PAIRS IN THE CRYSTAL STRUCTURE OF D(CGCGAATT(O8A)GCG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(A38)P*GP*CP*G)-3') | | Authors: | Leonard, G.A, Guy, A, Brown, T, Teoule, R, Hunter, W.N. | | Deposit date: | 1992-05-07 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformation of guanine-8-oxoadenine base pairs in the crystal structure of d(CGCGAATT(O8A)GCG).
Biochemistry, 31, 1992
|
|
1D58
 
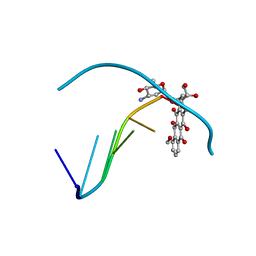 | | THE MOLECULAR STRUCTURE OF A 4'-EPIADRIAMYCIN COMPLEX WITH D(TGATCA) AT 1.7 ANGSTROM RESOLUTION-COMPARISON WITH THE STRUCTURE OF 4'-EPIADRIAMYCIN D(TGTACA) AND D(CGATCG) COMPLEXES | | Descriptor: | 4'-EPIDOXORUBICIN, DNA (5'-D(*TP*GP*AP*TP*CP*A)-3') | | Authors: | Langlois D'Estaintot, B, Gallois, B, Brown, T, Hunter, W.N. | | Deposit date: | 1992-02-20 | | Release date: | 1992-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular structure of a 4'-epiadriamycin complex with d(TGATCA) at 1.7A resolution: comparison with the structure of 4'-epiadriamycin d(TGTACA) and d(CGATCG) complexes.
Nucleic Acids Res., 20, 1992
|
|
6QJS
 
 | | Crystal structure of a DNA dodecamer containing a tetramethylpiperidinoxyl (nitroxide) spin label | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(P*GP*CP*AP*AP*AP*TP*TP*(S6M)P*GP*CP*G)-3') | | Authors: | Hardwick, J.S, Haugland, M.M, Ptchelkine, D, Brown, T, Anderson, E.E. | | Deposit date: | 2019-01-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 2'-Alkynyl spin-labelling is a minimally perturbing tool for DNA structural analysis.
Nucleic Acids Res., 48, 2020
|
|
1D67
 
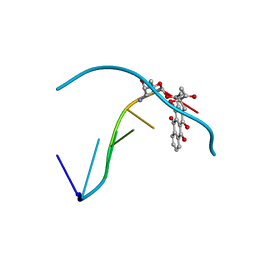 | | THE MOLECULAR STRUCTURE OF AN IDARUBICIN-D(TGATCA) COMPLEX AT HIGH RESOLUTION | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*CP*A)-3'), IDARUBICIN | | Authors: | Gallois, B, Langlois D'Estaintot, B, Brown, T, Hunter, W.N. | | Deposit date: | 1992-03-31 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of an idarubicin-d(TGATCA) complex at high resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1D54
 
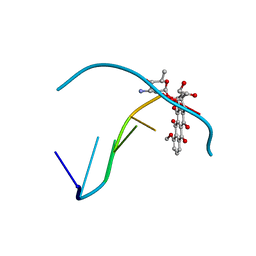 | |
1D90
 
 | | REFINED CRYSTAL STRUCTURE OF AN OCTANUCLEOTIDE DUPLEX WITH I.T MISMATCHED BASE PAIRS | | Descriptor: | DNA (5'-D(*GP*GP*IP*GP*CP*TP*CP*C)-3') | | Authors: | Cruse, W.B.T, Aymani, J, Kennard, O, Brown, T, Jack, A.G.C, Leonard, G.A. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined crystal structure of an octanucleotide duplex with I.T. mismatched base pairs.
Nucleic Acids Res., 17, 1989
|
|
1D27
 
 | |
1D97
 
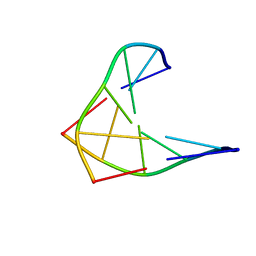 | | CHIRAL PHOSPHOROTHIOATE ANALOGUES OF B-DNA: THE CRYSTAL STRUCTURE OF RP-D(GP(S) CPGP(S)CPGP(S)C) | | Descriptor: | DNA (5'-D(RP*GP*(SC)P*GP*(SC)P*GP*(SC))-3') | | Authors: | Cruse, W.B.T, Salisbury, S.A, Brown, T, Cosstick, R, Eckstein, F, Kennard, O. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Chiral phosphorothioate analogues of B-DNA. The crystal structure of Rp-d[Gp(S)CpGp(S)CpGp(S)C].
J.Mol.Biol., 192, 1986
|
|
