6HPC
 
 | |
6HPB
 
 | |
5IQQ
 
 | |
4R71
 
 | | Structure of the Qbeta holoenzyme complex in the P1211 crystal form | | 分子名称: | 30S ribosomal protein S1, Elongation factor Ts, Elongation factor Tu, ... | | 著者 | Gytz, H, Seweryn, P, Kutlubaeva, Z, Chetverin, A.B, Brodersen, D.E, Knudsen, C.R. | | 登録日 | 2014-08-26 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.21 Å) | | 主引用文献 | Structural basis for RNA-genome recognition during bacteriophage Q beta replication.
Nucleic Acids Res., 43, 2015
|
|
6ZI1
 
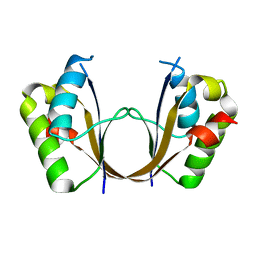 | | Crystal structure of the isolated H. influenzae VapD toxin (D7N mutant) | | 分子名称: | Endoribonuclease VapD | | 著者 | Bertelsen, M.B, Senissar, M, Nielsen, M.H, Bisiak, F, Cunha, M.V, Molinaro, A.L, Daines, D.A, Brodersen, D.E. | | 登録日 | 2020-06-24 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Basis for Toxin Inhibition in the VapXD Toxin-Antitoxin System.
Structure, 29, 2021
|
|
6ZI0
 
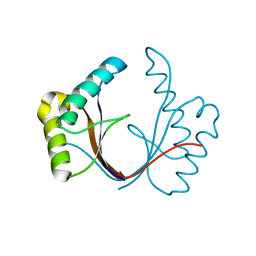 | | Crystal structure of the isolated H. influenzae VapD toxin (wildtype) | | 分子名称: | Endoribonuclease VapD | | 著者 | Bertelsen, M.B, Senissar, M, Nielsen, M.H, Bisiak, F, Cunha, M.V, Molinaro, A.L, Daines, D.A, Brodersen, D.E. | | 登録日 | 2020-06-24 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Basis for Toxin Inhibition in the VapXD Toxin-Antitoxin System.
Structure, 29, 2021
|
|
6ZN8
 
 | | Crystal structure of the H. influenzae VapXD toxin-antitoxin complex | | 分子名称: | Endoribonuclease VapD, VapX | | 著者 | Bertelsen, M.B, Senissar, M, Nielsen, M.H, Bisiak, F, Cunha, M.V, Molinaro, A.L, Daines, D.A, Brodersen, D.E. | | 登録日 | 2020-07-06 | | 公開日 | 2020-11-04 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.211 Å) | | 主引用文献 | Structural Basis for Toxin Inhibition in the VapXD Toxin-Antitoxin System.
Structure, 29, 2021
|
|
5K8J
 
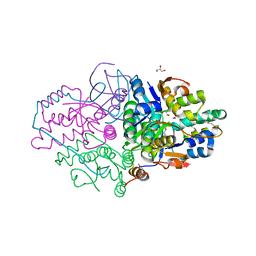 | | Structure of Caulobacter crescentus VapBC1 (apo form) | | 分子名称: | GLYCEROL, Ribonuclease VapC, VapB family protein | | 著者 | Bendtsen, K.L, Xu, K, Luckmann, M, Brodersen, D.E. | | 登録日 | 2016-05-30 | | 公開日 | 2016-12-28 | | 最終更新日 | 2018-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Toxin inhibition in C. crescentus VapBC1 is mediated by a flexible pseudo-palindromic protein motif and modulated by DNA binding.
Nucleic Acids Res., 45, 2017
|
|
7AB3
 
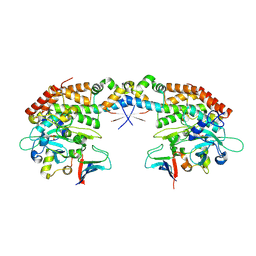 | |
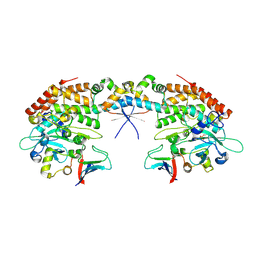
7AB5
 
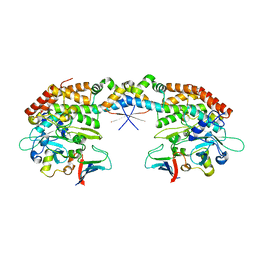 | |
7AB4
 
 | |
5L6L
 
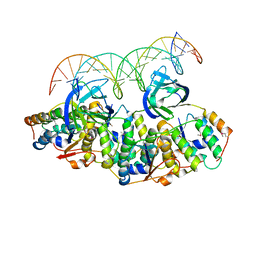 | | Structure of Caulobacter crescentus VapBC1 bound to operator DNA | | 分子名称: | DNA (27-MER), Ribonuclease VapC, VapB family protein | | 著者 | Bendtsen, K.L, Xu, K, Luckmann, M, Brodersen, D.E. | | 登録日 | 2016-05-30 | | 公開日 | 2016-12-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Toxin inhibition in C. crescentus VapBC1 is mediated by a flexible pseudo-palindromic protein motif and modulated by DNA binding.
Nucleic Acids Res., 45, 2017
|
|
5L6M
 
 | | Structure of Caulobacter crescentus VapBC1 (VapB1deltaC:VapC1 form) | | 分子名称: | GLYCEROL, MALONATE ION, Ribonuclease VapC, ... | | 著者 | Bendtsen, K.L, Xu, K, Luckmann, M, Brodersen, D.E. | | 登録日 | 2016-05-30 | | 公開日 | 2016-12-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Toxin inhibition in C. crescentus VapBC1 is mediated by a flexible pseudo-palindromic protein motif and modulated by DNA binding.
Nucleic Acids Res., 45, 2017
|
|
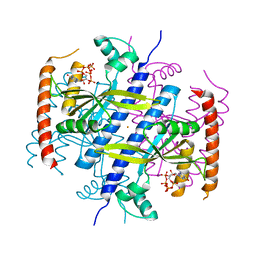
6EX0
 
 | |
6EWZ
 
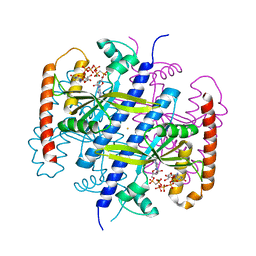 | |
6EXP
 
 | | Crystal structure of the SIRV3 AcrID1 (gp02) anti-CRISPR protein | | 分子名称: | SIRV3 AcrID1 (gp02) anti-CRISPR protein | | 著者 | He, F, Bhoobalan-Chitty, Y, Van, L.B, Kjeldsen, A.L, Dedola, M, Makarova, K.S, Koonin, E.V, Brodersen, D.E, Peng, X. | | 登録日 | 2017-11-08 | | 公開日 | 2018-01-31 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Anti-CRISPR proteins encoded by archaeal lytic viruses inhibit subtype I-D immunity.
Nat Microbiol, 3, 2018
|
|
6THH
 
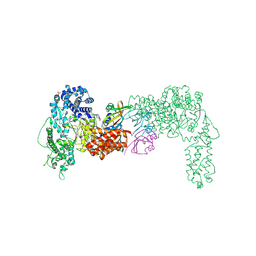 | |
4V7K
 
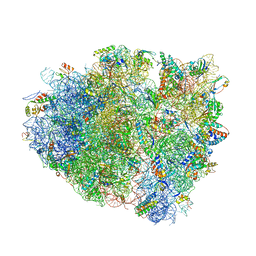 | | Structure of RelE nuclease bound to the 70S ribosome (postcleavage state) | | 分子名称: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | 著者 | Neubauer, C, Gao, Y.-G, Andersen, K.R, Dunham, C.M, Kelley, A.C, Hentschel, J, Gerdes, K, Ramakrishnan, V, Brodersen, D.E. | | 登録日 | 2009-11-02 | | 公開日 | 2014-07-09 | | 最終更新日 | 2014-12-10 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE.
Cell(Cambridge,Mass.), 139, 2009
|
|
4V4S
 
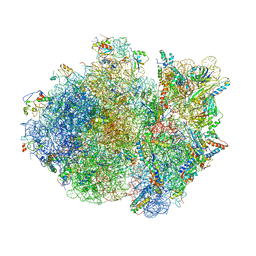 | | Crystal structure of the whole ribosomal complex. | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | 登録日 | 2005-10-12 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (6.76 Å) | | 主引用文献 | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
4V4T
 
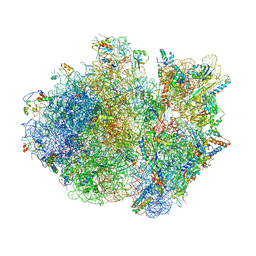 | | Crystal structure of the whole ribosomal complex with a stop codon in the A-site. | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | 登録日 | 2005-10-12 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (6.46 Å) | | 主引用文献 | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
5ED0
 
 | |
5ECY
 
 | |
5ECW
 
 | |
1HR0
 
 | | CRYSTAL STRUCTURE OF INITIATION FACTOR IF1 BOUND TO THE 30S RIBOSOMAL SUBUNIT | | 分子名称: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | 著者 | Carter, A.P, Clemons Jr, W.M, Brodersen, D.E, Morgan-Warren, R.J, Wimberly, B.T, Ramakrishnan, V. | | 登録日 | 2000-12-20 | | 公開日 | 2001-01-24 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal structure of an initiation factor bound to the 30S ribosomal subunit.
Science, 291, 2001
|
|
1IBK
 
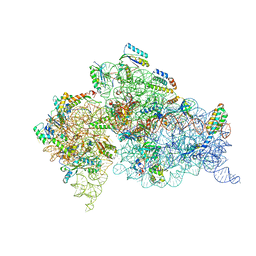 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH THE ANTIBIOTIC PAROMOMYCIN | | 分子名称: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | 著者 | Ogle, J.M, Brodersen, D.E, Clemons Jr, W.M, Tarry, M.J, Carter, A.P, Ramakrishnan, V. | | 登録日 | 2001-03-28 | | 公開日 | 2001-05-04 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | Recognition of cognate transfer RNA by the 30S ribosomal subunit.
Science, 292, 2001
|
|
