8PNY
 
 | | Crystal structure of D-amino acid aminotransferase from Blastococcus saxobsidens complexed with phenylhydrazine and in its apo form | | Descriptor: | Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase, [6-methyl-5-oxidanyl-4-[(2-phenylhydrazinyl)methyl]pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Expanded Substrate Specificity in D-Amino Acid Transaminases: A Case Study of Transaminase from Blastococcus saxobsidens.
Int J Mol Sci, 24, 2023
|
|
8PNW
 
 | | Crystal structure of D-amino acid aminotransferase from Blastococcus saxobsidens in holo form with PLP | | Descriptor: | Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase, CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Expanded Substrate Specificity in D-Amino Acid Transaminases: A Case Study of Transaminase from Blastococcus saxobsidens.
Int J Mol Sci, 24, 2023
|
|
5EEB
 
 | | Apo form of thermostable aldehyde dehydrogenase from Pyrobaculum sp. 1860 | | Descriptor: | Aldehyde dehydrogenase | | Authors: | Petrova, T.E, Bezsudnova, E.Y, Boyko, K.M, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-10-22 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.038 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
5EKC
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+ | | Descriptor: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Nikolaeva, A.Y, Rakitina, T.V, Shabalin, I.G, Popov, V.O. | | Deposit date: | 2015-11-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Structure of thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+
To Be Published
|
|
5EXF
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+ | | Descriptor: | Aldehyde dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Nikolaeva, A.Y, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2015-11-23 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
5F2C
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. 1860 crystallized in microgravity (complex with NADP+) | | Descriptor: | Aldehyde dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T.E, Bezsudnova, E.Y, Boyko, K.M, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-12-01 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
5EUY
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+ | | Descriptor: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Nikolaeva, A.Y, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2015-11-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
7P8O
 
 | | Crystal structure of D-aminoacid transaminase from Haliscomenobacter hydrossis in its intermediate form | | Descriptor: | Aminotransferase class IV, MAGNESIUM ION, SULFATE ION | | Authors: | Matyuta, I.O, Boyko, K.M, Bakunova, A.K, Nikolaeva, A.Y, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2021-07-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of D-aminoacid transaminase from Haliscomenobacter hydrossis in its apo form
To Be Published
|
|
6QVM
 
 | | Undecaheme cytochrome from S-layer of Carboxydothermus ferrireducens | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Osipov, E.M, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Gavrilov, S.F, Popov, V.O. | | Deposit date: | 2019-03-04 | | Release date: | 2020-03-25 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Extracellular Fe(III) reductase structure reveals a modular organization enabling S-layer insertion and electron transfer to insoluble substrates
Structure, 2023
|
|
4NMK
 
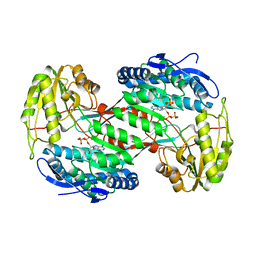 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. crystallized in microgravity (complex with NADP+) | | Descriptor: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Boyko, K.M, Bezsudnova, E.Y, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | Deposit date: | 2013-11-15 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. crystallized in microgravity (complex with NADP+)
To be Published
|
|
4NMJ
 
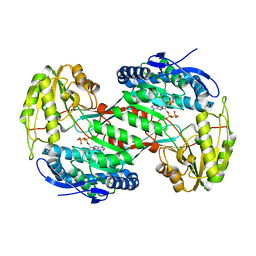 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. complexed with NADP+ at 2 A resolution | | Descriptor: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Boyko, K.M, Bezsudnova, E.Y, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | Deposit date: | 2013-11-15 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. complexed with NADP+ at 2 A resolution
To be Published
|
|
8P3L
 
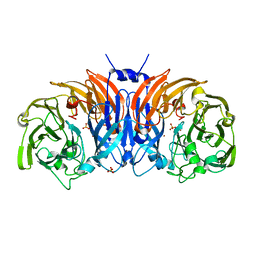 | | The structure of thiocyanate dehydrogenase mutant form with Thr 169 replaced by Ala from Thioalkalivibrio paradoxus | | Descriptor: | COPPER (II) ION, SULFATE ION, Twin-arginine translocation signal domain-containing protein | | Authors: | Varfolomeeva, L.A, Polyakov, K.M, Komolov, A.S, Rakitina, T.V, Dergousova, N.I, Dorovatovskii, P.V, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2023-05-18 | | Release date: | 2023-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Improvement of the Diffraction Properties of Thiocyanate Dehydrogenase Crystals
Crystallography Reports, 2023
|
|
8P3M
 
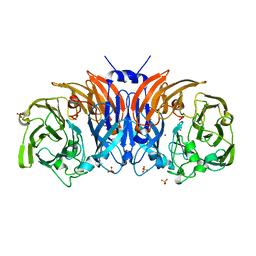 | | The structure of thiocyanate dehydrogenase mutant form with Lys 281 replaced by Ala from Thioalkalivibrio paradoxus | | Descriptor: | BORIC ACID, COPPER (II) ION, SODIUM ION, ... | | Authors: | Varfolomeeva, L.A, Polyakov, K.M, Komolov, A.S, Rakitina, T.V, Dergousova, N.I, Dorovatovskii, P.V, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2023-05-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Improvement of the Diffraction Properties of Thiocyanate Dehydrogenase Crystals
Crystallography Reports, 2023
|
|
8Q9Y
 
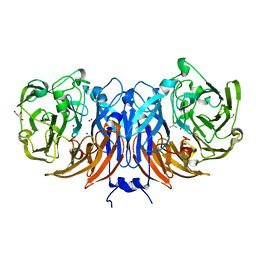 | | The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum in complex with inhibitor thiourea at 1.10 A resolution | | Descriptor: | COPPER (II) ION, GLYCEROL, THIOUREA, ... | | Authors: | Varfolomeeva, L.A, Polyakov, K.M, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2023-08-22 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum at atomic resolution
To Be Published
|
|
8Q9X
 
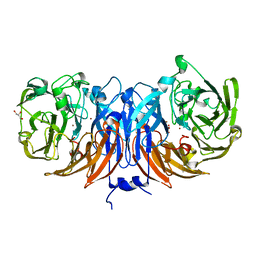 | | The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum with molecular oxygen at 1.05 A resolution | | Descriptor: | COPPER (II) ION, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | Varfolomeeva, L.A, Polyakov, K.M, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2023-08-22 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum at atomic resolution
To Be Published
|
|
6YTS
 
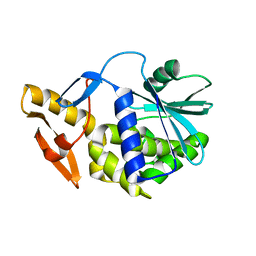 | | Solution NMR structure of type-I ribosome-inactivating protein trichobakin (TBK) | | Descriptor: | Trichobakin | | Authors: | Britikov, V.V, Bocharov, E.V, Britikova, E.V, Le, T.B.T, Phan, C.V, Boyko, K.M, Arseniev, A.S, Usanov, S.A. | | Deposit date: | 2020-04-24 | | Release date: | 2020-05-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of type-I ribosome-inactivating protein trichobakin (TBK)
To Be Published
|
|
6ZL7
 
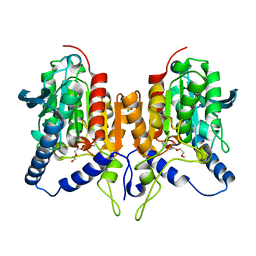 | | CRYSTAL STRUCTURE OF C173S MUTATION IN THE PMGL2 ESTERASE FROM PERMAFROST METAGENOMIC LIBRARY | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, MAGNESIUM ION, PMGL2 | | Authors: | Goryaynova, D.A, Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Kryukova, M.V, Petrovskaya, L.E, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Popov, V.O. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF C173S MUTATION IN THE PMGL2 ESTERASE FROM PERMAFROST METAGENOMIC LIBRARY
To Be Published
|
|
8AHR
 
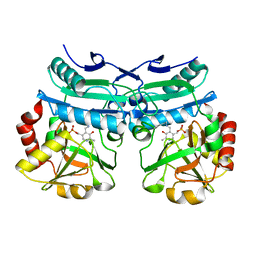 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense in holo form with PLP | | Descriptor: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-07-22 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense.
Molecules, 28, 2023
|
|
8AHU
 
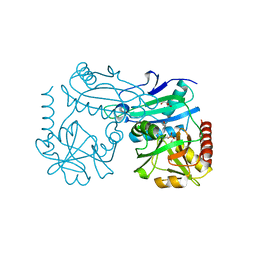 | | Crystal structure of D-amino acid aminotrensferase from Haliscomenobacter hydrossis complexed with D-cycloserine | | Descriptor: | Aminotransferase class IV, GLYCEROL, [5-hydroxy-6-methyl-4-({[(4E)-3-oxo-1,2-oxazolidin-4-ylidene]amino}methyl)pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Bakunova, A.K, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-07-22 | | Release date: | 2022-08-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Mechanism of D-Cycloserine Inhibition of D-Amino Acid Transaminase from Haliscomenobacter hydrossis.
Biochemistry Mosc., 88, 2023
|
|
7ZVQ
 
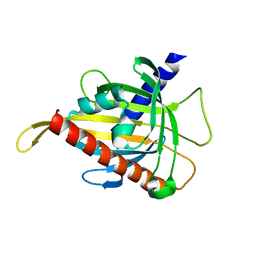 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform, S206V mutant | | Descriptor: | Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the carotenoid binding and transport function of a START domain.
Structure, 30, 2022
|
|
7ZTR
 
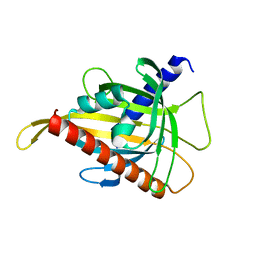 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform, W232F mutant | | Descriptor: | Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Silkworm carotenoprotein as an efficient carotenoid extractor, solubilizer and transporter.
Int.J.Biol.Macromol., 223, 2022
|
|
7ZTU
 
 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform, D162L mutant | | Descriptor: | Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Silkworm carotenoprotein as an efficient carotenoid extractor, solubilizer and transporter.
Int.J.Biol.Macromol., 223, 2022
|
|
7ZVR
 
 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) complexed with zeaxanthin | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the carotenoid binding and transport function of a START domain.
Structure, 30, 2022
|
|
7ZTQ
 
 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform | | Descriptor: | Carotenoid-binding protein, GLYCEROL | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Silkworm carotenoprotein as an efficient carotenoid extractor, solubilizer and transporter.
Int.J.Biol.Macromol., 223, 2022
|
|
8AIE
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense complexed with D-cycloserine | | Descriptor: | 3-azanyloxy-2-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]propanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class IV, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3D Structure of D-Аmino Acid Тransaminase from Aminobacterium colombiense in Complex with D-Cycloserine
Crystallography Reports, 68, 2023
|
|
