4X6H
 
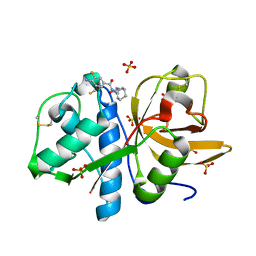 | | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors. | | 分子名称: | 4-amino-3-fluoro-N-(1-{[(2Z)-2-iminoethyl]carbamoyl}cyclohexyl)benzamide, 4-amino-N-{1-[(cyanomethyl)carbamoyl]cyclohexyl}-3-fluorobenzamide, Cathepsin K, ... | | 著者 | Borisek, J, Mohar, B, Vizovisek, M, Sosnowski, P, Turk, D, Turk, B, Novic, M. | | 登録日 | 2014-12-08 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors.
J.Med.Chem., 58, 2015
|
|
4X6J
 
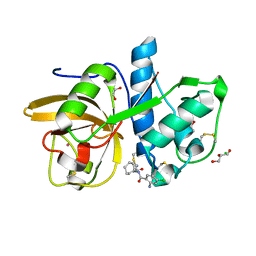 | | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors. | | 分子名称: | 2-amino-4-chloro-N-(1-{[(2E)-2-iminoethyl]carbamoyl}cyclohexyl)benzamide, CHLORIDE ION, Cathepsin K, ... | | 著者 | Borisek, J, Mohar, B, Vizovisek, M, Sosnowski, P, Turk, D, Turk, B, Novic, M. | | 登録日 | 2014-12-08 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors.
J.Med.Chem., 58, 2015
|
|
4X6I
 
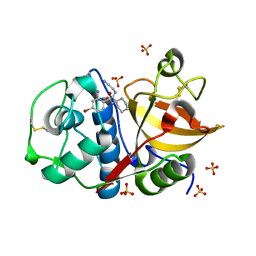 | | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors. | | 分子名称: | 2-amino-4-bromo-N-{1-[(cyanomethyl)carbamoyl]cyclohexyl}benzamide, Cathepsin K, SULFATE ION | | 著者 | Borisek, J, Mohar, B, Vizovisek, M, Sosnowski, P, Turk, D, Turk, B, Novic, M. | | 登録日 | 2014-12-08 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors.
J.Med.Chem., 58, 2015
|
|
7JQL
 
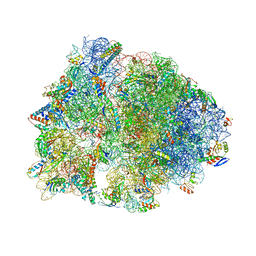 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Bac7-001, mRNA, and deacylated P-site tRNA at 3.00A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Mardirossian, M, Sola, R, Beckert, B, Valencic, E, Collis, D.W.P, Borisek, J, Armas, F, Di Stasi, A, Buchmann, J, Syroegin, E.A, Polikanov, Y.S, Magistrato, A, Hilpert, K, Wilson, D.N, Scocchi, M. | | 登録日 | 2020-08-11 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Peptide Inhibitors of Bacterial Protein Synthesis with Broad Spectrum and SbmA-Independent Bactericidal Activity against Clinical Pathogens.
J.Med.Chem., 63, 2020
|
|
7JQM
 
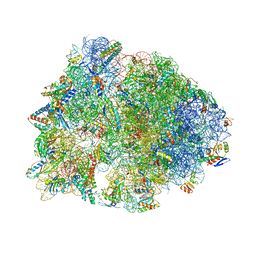 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Bac7-002, mRNA, and deacylated P-site tRNA at 3.05A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Mardirossian, M, Sola, R, Beckert, B, Valencic, E, Collis, D.W.P, Borisek, J, Armas, F, Di Stasi, A, Buchmann, J, Syroegin, E.A, Polikanov, Y.S, Magistrato, A, Hilpert, K, Wilson, D.N, Scocchi, M. | | 登録日 | 2020-08-11 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Peptide Inhibitors of Bacterial Protein Synthesis with Broad Spectrum and SbmA-Independent Bactericidal Activity against Clinical Pathogens.
J.Med.Chem., 63, 2020
|
|
5J6Q
 
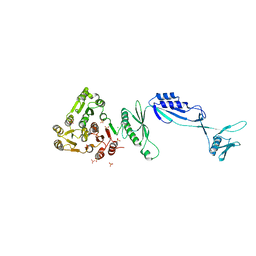 | | Cwp8 from Clostridium difficile | | 分子名称: | CHLORIDE ION, Cell wall binding protein cwp8, SULFATE ION | | 著者 | Renko, M, Usenik, A, Turk, D. | | 登録日 | 2016-04-05 | | 公開日 | 2017-02-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The CWB2 Cell Wall-Anchoring Module Is Revealed by the Crystal Structures of the Clostridium difficile Cell Wall Proteins Cwp8 and Cwp6.
Structure, 25, 2017
|
|
6QBE
 
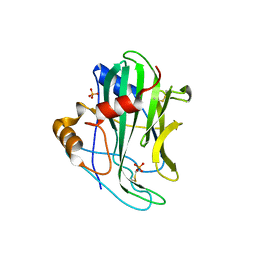 | | Crystal structure of non-toxic HaNLP3 protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nep1-like protein, PHOSPHATE ION, ... | | 著者 | Lenarcic, T, Podobnik, M, Anderluh, G. | | 登録日 | 2018-12-21 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular basis for functional diversity among microbial Nep1-like proteins.
Plos Pathog., 15, 2019
|
|
6QBD
 
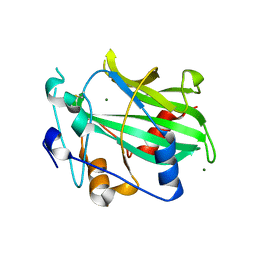 | | Crystal structure of NLPPya P41A, D44N, N48E mutant | | 分子名称: | 25 kDa protein elicitor, MAGNESIUM ION | | 著者 | Lenarcic, T, Podobnik, M, Anderluh, G. | | 登録日 | 2018-12-21 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Molecular basis for functional diversity among microbial Nep1-like proteins.
Plos Pathog., 15, 2019
|
|
6FXO
 
 | | Crystal structure of Major Bifunctional Autolysin | | 分子名称: | Bifunctional autolysin, CHLORIDE ION | | 著者 | Pintar, S, Turk, D. | | 登録日 | 2018-03-09 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Domain sliding of two Staphylococcus aureus N-acetylglucosaminidases enables their substrate-binding prior to its catalysis.
Commun Biol, 3, 2020
|
|
6FXP
 
 | | Crystal structure of S. aureus glucosaminidase B | | 分子名称: | CHLORIDE ION, SODIUM ION, TETRAETHYLENE GLYCOL, ... | | 著者 | Pintar, S, Turk, D. | | 登録日 | 2018-03-09 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Domain sliding of two Staphylococcus aureus N-acetylglucosaminidases enables their substrate-binding prior to its catalysis.
Commun Biol, 3, 2020
|
|
5J72
 
 | | Cwp6 from Clostridium difficile | | 分子名称: | CALCIUM ION, CHLORIDE ION, CITRIC ACID, ... | | 著者 | Renko, M, Usenik, A, Turk, D. | | 登録日 | 2016-04-05 | | 公開日 | 2017-02-08 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The CWB2 Cell Wall-Anchoring Module Is Revealed by the Crystal Structures of the Clostridium difficile Cell Wall Proteins Cwp8 and Cwp6.
Structure, 25, 2017
|
|
