7QCO
 
 | | The structure of Photosystem I tetramer from Chroococcidiopsis TS-821, a thermophilic, unicellular, non-heterocyst-forming cyanobacterium | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Semchonok, D.A, Mondal, J, Cooper, J.C, Schlum, K, Li, M, Amin, M, Sorzano, C.O.S, Ramirez-Aportela, E, Kastritis, P.L, Boekema, E.J, Guskov, A, Bruce, B.D. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a tetrameric photosystem I from Chroococcidiopsis TS-821, a thermophilic, unicellular, non-heterocyst-forming cyanobacterium.
Plant Commun., 3, 2022
|
|
5MYJ
 
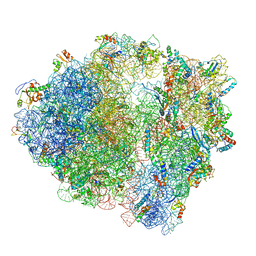 | | Structure of 70S ribosome from Lactococcus lactis | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Franken, L.E, Oostergetel, G.T, Pijning, T, Puri, P, Boekema, E.J, Poolman, B, Guskov, A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-10-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | A general mechanism of ribosome dimerization revealed by single-particle cryo-electron microscopy.
Nat Commun, 8, 2017
|
|
5MDX
 
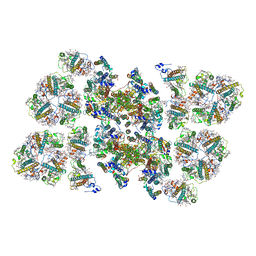 | | Cryo-EM structure of the PSII supercomplex from Arabidopsis thaliana | | Descriptor: | CHLOROPHYLL A, CHLOROPHYLL B, Chlorophyll a-b binding protein 1, ... | | Authors: | van Bezouwen, L.S, Caffarri, S, Kale, R.S, Kouril, R, Thunnissen, A.M.W.H, Oostergetel, G.T, Boekema, E.J. | | Deposit date: | 2016-11-13 | | Release date: | 2017-06-21 | | Last modified: | 2019-04-24 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Subunit and chlorophyll organization of the plant photosystem II supercomplex.
Nat Plants, 3, 2017
|
|
3O20
 
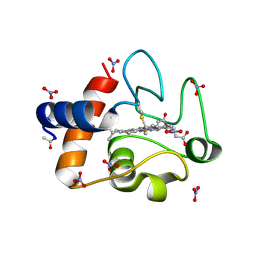 | | Electron transfer complexes:experimental mapping of the Redox-dependent Cytochrome C electrostatic surface | | Descriptor: | Cytochrome c, HEME C, NITRATE ION | | Authors: | De March, M, De Zorzi, R, Casini, A, Messori, L, Geremia, S, Demitri, N, Gabbiani, C, Guerri, A. | | Deposit date: | 2010-07-22 | | Release date: | 2012-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nitrate as a probe of cytochrome c surface: crystallographic identification of crucial "hot spots" for protein-protein recognition.
J. Inorg. Biochem., 135, 2014
|
|
3O1Y
 
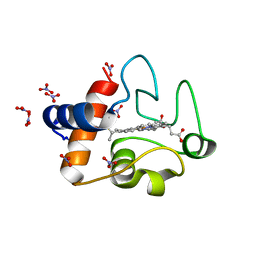 | | Electron transfer complexes: Experimental mapping of the redox-dependent cytochrome c electrostatic surface | | Descriptor: | Cytochrome c, HEME C, NITRATE ION | | Authors: | De March, M, De Zorzi, R, Demitri, N, Gabbiani, C, Guerri, A, Casini, A, Messori, L, Geremia, S. | | Deposit date: | 2010-07-22 | | Release date: | 2012-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Nitrate as a probe of cytochrome c surface: crystallographic identification of crucial "hot spots" for protein-protein recognition.
J. Inorg. Biochem., 135, 2014
|
|
