7PHQ
 
 | | Structure of homo-dimeric Staphylococcus capitis divalent metal ion transporter (DMT) by NabFab-fiducial assisted cryo-EM | | Descriptor: | Anti-Fab nanobody, DMT-Nb16_4, Divalent metal cation transporter MntH, ... | | Authors: | Bloch, J.S, Mukherjee, S, Kowal, J, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2021-08-18 | | Release date: | 2021-09-01 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (8.45 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7PIJ
 
 | | Structure of Staphylococcus capitis divalent metal ion transporter (DMT) by NabFab-fiducial assisted cryo-EM | | Descriptor: | Anti-Fab nanobody, DMTNb16_4, Divalent metal cation transporter MntH, ... | | Authors: | Bloch, J.S, Mukherjee, S, Kowal, J, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2021-08-20 | | Release date: | 2021-09-01 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7PHP
 
 | | Structure of Multidrug and Toxin Compound Extrusion (MATE) transporter NorM by NabFab-fiducial assisted cryo-EM | | Descriptor: | Anti-Fab nanobody, Multidrug resistance protein NorM, NabFab HC, ... | | Authors: | Bloch, J.S, Mukherjee, S, Kowal, J, Niederer, M, Pardon, E, Steyaert, J, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2021-08-18 | | Release date: | 2021-09-01 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5NSA
 
 | | Beta domain of human transcobalamin bound to Co-beta-[2-(2,4-difluorophenyl)ethinyl]cobalamin | | Descriptor: | 1-ethynyl-2,4-difluorobenzene, CALCIUM ION, COBALAMIN, ... | | Authors: | Bloch, J.S, Ruetz, M, Krautler, B, Locher, K.P. | | Deposit date: | 2017-04-25 | | Release date: | 2017-09-27 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.273 Å) | | Cite: | Structure of the human transcobalamin beta domain in four distinct states.
PLoS ONE, 12, 2017
|
|
7QBG
 
 | | TC:CD320 in complex with nanobody TC-Nb4 | | Descriptor: | Anti-TC:CD320 nanobody TC-Nb4, CALCIUM ION, CD320 antigen, ... | | Authors: | Bloch, J.S, Locher, K.P. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Generation of nanobodies targeting the human, transcobalamin-mediated vitamin B 12 uptake route.
Faseb J., 36, 2022
|
|
7QBE
 
 | | TC:CD320 in complex with nanobody TC-Nb11 | | Descriptor: | Anti-transcobalamin-2 nanobody TC-Nb11, CALCIUM ION, CD320 antigen, ... | | Authors: | Bloch, J.S, Locher, K.P. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Generation of nanobodies targeting the human, transcobalamin-mediated vitamin B 12 uptake route.
Faseb J., 36, 2022
|
|
7QBF
 
 | | TC:CD320 in complex with nanobody TC-Nb34 | | Descriptor: | 1,2-ETHANEDIOL, Anti-transcobalamin-2 nanobody TC-Nb34, CALCIUM ION, ... | | Authors: | Bloch, J.S, Locher, K.P. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Generation of nanobodies targeting the human, transcobalamin-mediated vitamin B 12 uptake route.
Faseb J., 36, 2022
|
|
7QBD
 
 | | TC:CD320 in complex with nanobody TC-Nb26 | | Descriptor: | Antitranscobalamin-nanobody TC-Nb26, CALCIUM ION, CD320 antigen, ... | | Authors: | Bloch, J.S, Locher, K.P. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.18 Å) | | Cite: | Generation of nanobodies targeting the human, transcobalamin-mediated vitamin B 12 uptake route.
Faseb J., 36, 2022
|
|
6SNI
 
 | | Cryo-EM structure of nanodisc reconstituted yeast ALG6 in complex with 6AG9 Fab | | Descriptor: | 6AG9-Fab heavy chain, 6AG9-Fab light chain, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Bloch, J.S, Pesciullesi, G, Boilevin, J, Nosol, K, Irobalieva, R.N, Darbre, T, Aebi, M, Kossiakoff, A.A, Reymond, J.L, Locher, K.P. | | Deposit date: | 2019-08-24 | | Release date: | 2020-03-11 | | Last modified: | 2020-04-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and mechanism of the ER-based glucosyltransferase ALG6.
Nature, 579, 2020
|
|
6SNH
 
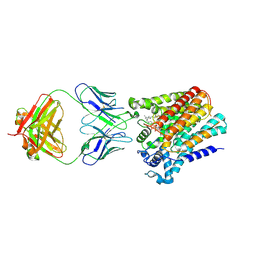 | | Cryo-EM structure of yeast ALG6 in complex with 6AG9 Fab and Dol25-P-Glc | | Descriptor: | 6AG9 Fab heavy chain, 6AG9 Fab light chain, Dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase, ... | | Authors: | Bloch, J.S, Pesciullesi, G, Boilevin, J, Nosol, K, Irobalieva, R.N, Darbre, T, Aebi, M, Kossiakoff, A.A, Reymond, J.L, Locher, K.P. | | Deposit date: | 2019-08-24 | | Release date: | 2020-03-11 | | Last modified: | 2020-04-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and mechanism of the ER-based glucosyltransferase ALG6.
Nature, 579, 2020
|
|
7ZLH
 
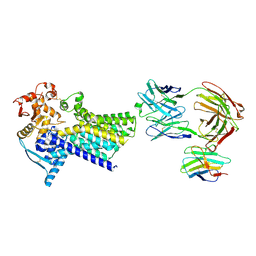 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in apo state, bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Irobalieva, R, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZLI
 
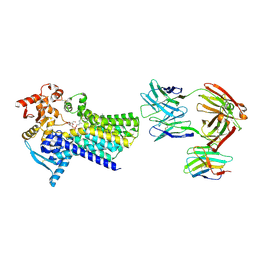 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with Dol25-P-Man and bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZLG
 
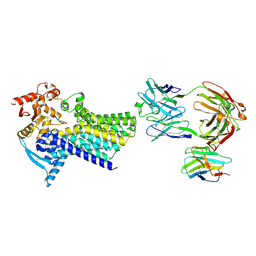 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with acceptor peptide and bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Mao, R, Irobalieva, R, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZLJ
 
 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in ternary complex with Dol25-P-C-Man and acceptor peptide, bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mao, R, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
5NO0
 
 | |
5NRP
 
 | |
5NP4
 
 | |
7RTH
 
 | | Crystal structure of an anti-lysozyme nanobody in complex with an anti-nanobody Fab "NabFab" | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Fragment Antigen-Binding Heavy Chain, ... | | Authors: | Filippova, E.V, Mukherjee, S, Bloch, J.S, Locher, K.P, Kossiakoff, A.A. | | Deposit date: | 2021-08-13 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6SRW
 
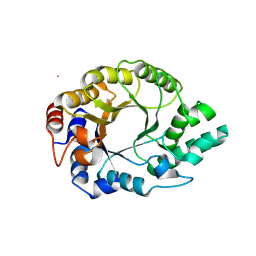 | | Kemp Eliminase HG3.17 mutant Q50F, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, GLYCEROL, Kemp Eliminase HG3.17 Q50F | | Authors: | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
6SRZ
 
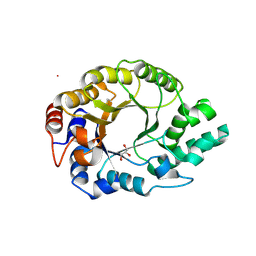 | | Kemp Eliminase HG3.17 mutant Q50H, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, GLYCEROL, Kemp Eliminase HG3.17 Q50H, ... | | Authors: | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
6SS1
 
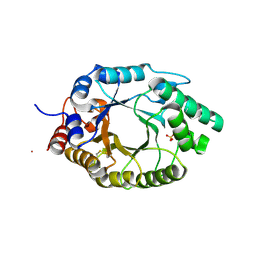 | | Kemp Eliminase HG3.17 mutant Q50A, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3.17 Q50A, E47N,N300D Complexed with Transition State Analog 6-Nitrobenzotriazole, ... | | Authors: | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
6SS3
 
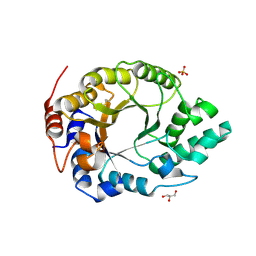 | | Kemp Eliminase HG3.17 mutant Q50K, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, GLYCEROL, Kemp Eliminase HG3.17 Q50K, ... | | Authors: | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
6SRY
 
 | | Kemp Eliminase HG3.17 mutant Q50S, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3.17 Q50S, E47N,N300D, ... | | Authors: | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
6TU6
 
 | |
8AGB
 
 | | Structure of yeast oligosaccharylransferase complex with lipid-linked oligosaccharide bound | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ramirez, A.S, de Capitani, M, Pesciullesi, G, Kowal, J, Bloch, J.S, Irobalieva, R.N, Aebi, M, Reymond, J.L, Locher, K.P. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for glycan recognition and reaction priming of eukaryotic oligosaccharyltransferase.
Nat Commun, 13, 2022
|
|
