1GKS
 
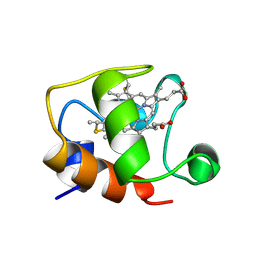 | | ECTOTHIORHODOSPIRA HALOPHILA CYTOCHROME C551 (REDUCED), NMR, 37 STRUCTURES | | Descriptor: | CYTOCHROME C551, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bersch, B, Blackledge, M.J, Meyer, T.E, Marion, D. | | Deposit date: | 1996-07-16 | | Release date: | 1997-01-27 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Ectothiorhodospira halophila ferrocytochrome c551: solution structure and comparison with bacterial cytochromes c.
J.Mol.Biol., 264, 1996
|
|
1APQ
 
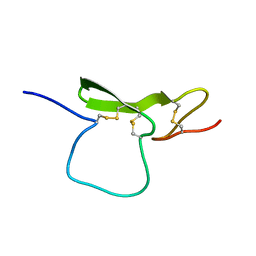 | | STRUCTURE OF THE EGF-LIKE MODULE OF HUMAN C1R, NMR, 19 STRUCTURES | | Descriptor: | COMPLEMENT PROTEASE C1R | | Authors: | Bersch, B, Hernandez, J.-F, Marion, D, Arlaud, G.J. | | Deposit date: | 1997-07-22 | | Release date: | 1997-09-17 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the epidermal growth factor (EGF)-like module of human complement protease C1r, an atypical member of the EGF family.
Biochemistry, 37, 1998
|
|
3ZFJ
 
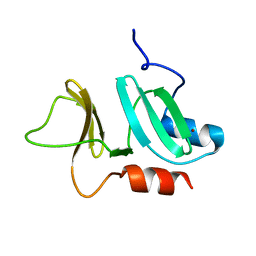 | | N-terminal domain of pneumococcal PhtD protein with bound Zn(II) | | Descriptor: | PNEUMOCOCCAL HISTIDINE TRIAD PROTEIN D, ZINC ION | | Authors: | Bersch, B, Bougault, C, Favier, A, Gabel, F, Roux, L, Vernet, T, Durmort, C. | | Deposit date: | 2012-12-11 | | Release date: | 2013-11-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | New Insights Into Histidine Triad Proteins: Solution Structure of a Streptococcus Pneumoniae Phtd Domain and Zinc Transfer to Adcaii.
Plos One, 8, 2013
|
|
6ZXP
 
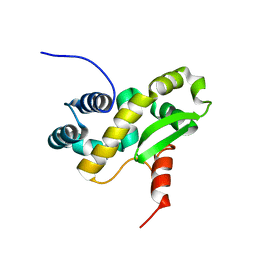 | | Solution structure of the C-terminal domain of the vaccinia virus DNA polymerase processivity factor component A20 fused to a short peptide from the viral DNA polymerase E9. | | Descriptor: | DNA polymerase processivity factor component A20,DNA polymerase processivity factor component E9 | | Authors: | Bersch, B, Tarbouriech, N, Burmeister, W, Iseni, F. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Domain of A20, the Missing Brick for the Characterization of the Interface between Vaccinia Virus DNA Polymerase and its Processivity Factor.
J.Mol.Biol., 433, 2021
|
|
6ZYC
 
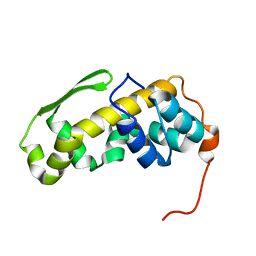 | | Solution structure of the C-terminal domain of the vaccinia virus DNA polymerase processivity factor component A20. | | Descriptor: | DNA polymerase processivity factor component A20 | | Authors: | Bersch, B, Iseni, F, Burmeister, W, Tarbouriech, N. | | Deposit date: | 2020-07-31 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Domain of A20, the Missing Brick for the Characterization of the Interface between Vaccinia Virus DNA Polymerase and its Processivity Factor.
J.Mol.Biol., 433, 2021
|
|
2K0Q
 
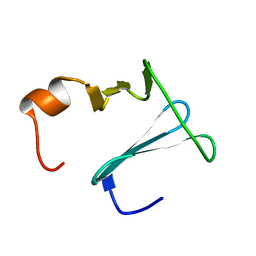 | | Solution structure of CopK, a periplasmic protein involved in copper resistance in Cupriavidus metallidurans CH34 | | Descriptor: | Putative uncharacterized protein copK | | Authors: | Bersch, B, Favier, A, Schanda, P, Coves, J, van Aelst, S, Vallaeys, T, Wattiez, R, Mergeay, M. | | Deposit date: | 2008-02-12 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Molecular structure and metal-binding properties of the periplasmic CopK protein expressed in Cupriavidus metallidurans CH34 during copper challenge.
J.Mol.Biol., 380, 2008
|
|
2L55
 
 | |
5A4G
 
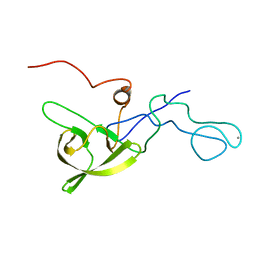 | | NMR structure of a 180 residue construct encompassing the N-terminal metal-binding site and the membrane proximal domain of SilB from Cupriavidus metallidurans CH34 | | Descriptor: | SILB, SILVER EFFLUX PROTEIN, MFP COMPONENT OF THE THREE COMPONENTS PROTON ANTIPORTER METAL EFFLUX SYSTEM, ... | | Authors: | Bersch, B, Urbina Fernandez, P, Vandenbussche, G. | | Deposit date: | 2015-06-09 | | Release date: | 2016-05-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Investigation of the Ag+/Cu+-Binding Domains of the Periplasmic Adaptor Protein Silb from Cupriavidus Metallidurans Ch34.
Biochemistry, 55, 2016
|
|
2KM0
 
 | | Cu(I)-bound CopK | | Descriptor: | COPPER (I) ION, Copper resistance protein K | | Authors: | Bersch, B. | | Deposit date: | 2009-07-15 | | Release date: | 2010-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | CopK from Cupriavidus metallidurans CH34 Binds Cu(I) in a Tetrathioether Site: Characterization by X-ray Absorption and NMR Spectroscopy
J.Am.Chem.Soc., 2010
|
|
1YKG
 
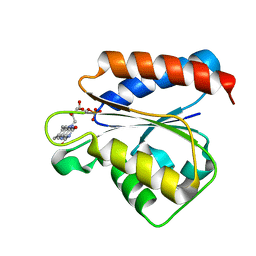 | | Solution structure of the flavodoxin-like domain from the Escherichia coli sulfite reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, Sulfite reductase [NADPH] flavoprotein alpha-component | | Authors: | Sibille, N, Blackledge, M, Brutscher, B, Coves, J, Bersch, B. | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Sulfite Reductase Flavodoxin-like Domain from Escherichia coli
Biochemistry, 44, 2005
|
|
2FU4
 
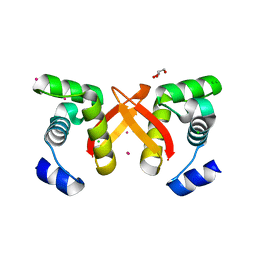 | | Crystal Structure of the DNA binding domain of E.coli FUR (Ferric Uptake Regulator) | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferric uptake regulation protein, ... | | Authors: | Pecqueur, L, D'Autreaux, B, Dupuy, J, Nicolet, Y, Jacquamet, L, Brutscher, B, Michaud-Soret, I, Bersch, B. | | Deposit date: | 2006-01-26 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural changes of Escherichia coli ferric uptake regulator during metal-dependent dimerization and activation explored by NMR and X-ray crystallography
J.Biol.Chem., 281, 2006
|
|
2LRS
 
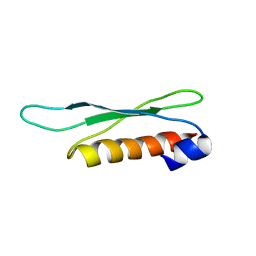 | | The second dsRBD domain from A. thaliana DICER-LIKE 1 | | Descriptor: | Endoribonuclease Dicer homolog 1 | | Authors: | Burdisso, P, Suarez, I, Bersch, B, Bologna, N, Palatnik, J, Boisbouvier, J, Rasia, R. | | Deposit date: | 2012-04-12 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Second Double-Stranded RNA Binding Domain of Dicer-like Ribonuclease 1: Structural and Biochemical Characterization.
Biochemistry, 51, 2012
|
|
7N82
 
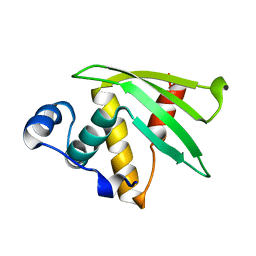 | | NMR Solution structure of Se0862 | | Descriptor: | Biofilm-related protein | | Authors: | Zhang, N, LiWang, A.L. | | Deposit date: | 2021-06-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Assessment of prediction methods for protein structures determined by NMR in CASP14: Impact of AlphaFold2.
Proteins, 89, 2021
|
|
4H2D
 
 | | Crystal structure of NDOR1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH-dependent diflavin oxidoreductase 1 | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mikolajczyk, M, Jaiswal, D, Winkelmann, J. | | Deposit date: | 2012-09-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular view of an electron transfer process essential for iron-sulfur protein biogenesis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
