4W8W
 
 | |
4W8Z
 
 | |
4W8V
 
 | |
4W8Y
 
 | | Structure of full length Cmr2 from Pyrococcus furiosus (Manganese bound form) | | 分子名称: | CRISPR system Cmr subunit Cmr2, MANGANESE (II) ION, ZINC ION | | 著者 | Benda, C, Ebert, J, Baumgaertner, M, Conti, E. | | 登録日 | 2014-08-26 | | 公開日 | 2014-10-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Model of a CRISPR RNA-Silencing Complex Reveals the RNA-Target Cleavage Activity in Cmr4.
Mol.Cell, 56, 2014
|
|
4W8X
 
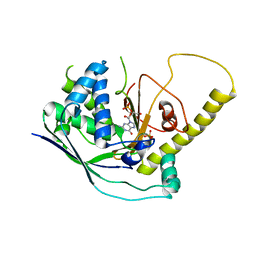 | | Crystal Structure of Cmr1 from Pyrococcus furiosus bound to a nucleotide | | 分子名称: | CRISPR system Cmr subunit Cmr1-1, GUANOSINE-3'-MONOPHOSPHATE, PHOSPHATE ION | | 著者 | Benda, C, Ebert, J, Baumgaertner, M, Conti, E. | | 登録日 | 2014-08-26 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Model of a CRISPR RNA-Silencing Complex Reveals the RNA-Target Cleavage Activity in Cmr4.
Mol.Cell, 56, 2014
|
|
1K68
 
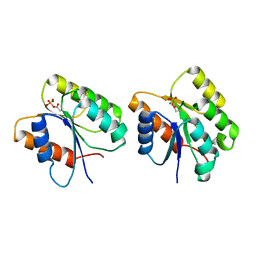 | | Crystal Structure of the Phosphorylated Cyanobacterial Phytochrome Response Regulator RcpA | | 分子名称: | MAGNESIUM ION, Phytochrome Response Regulator RcpA | | 著者 | Benda, C, Scheufler, C, Tandeau de Marsac, N, Gaertner, W. | | 登録日 | 2001-10-15 | | 公開日 | 2003-12-16 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of two cyanobacterial response regulators in apo- and phosphorylated form reveal a novel dimerization motif of phytochrome-associated response regulators
Biophys.J., 87, 2004
|
|
1K66
 
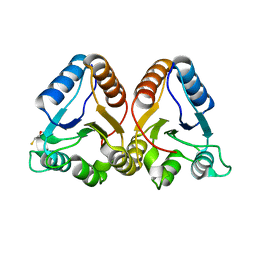 | | Crystal Structure of the Cyanobacterial Phytochrome Response Regulator, RcpB | | 分子名称: | BETA-MERCAPTOETHANOL, Phytochrome Response Regulator RcpB | | 著者 | Benda, C, Scheufler, C, Tandeau de Marsac, N, Gaertner, W. | | 登録日 | 2001-10-15 | | 公開日 | 2003-12-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structures of two cyanobacterial response regulators in apo- and phosphorylated form reveal a novel dimerization motif of phytochrome-associated response regulators
Biophys.J., 87, 2004
|
|
7AT8
 
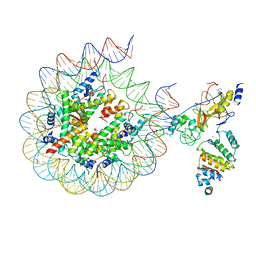 | | Histone H3 recognition by nucleosome-bound PRC2 subunit EZH2. | | 分子名称: | Histone H2A, Histone H2B 1.1, Histone H3.2, ... | | 著者 | Finogenova, K, Benda, C, Schaefer, I.B, Poepsel, S, Strauss, M, Mueller, J. | | 登録日 | 2020-10-29 | | 公開日 | 2020-12-09 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structural basis for PRC2 decoding of active histone methylation marks H3K36me2/3.
Elife, 9, 2020
|
|
5J8Y
 
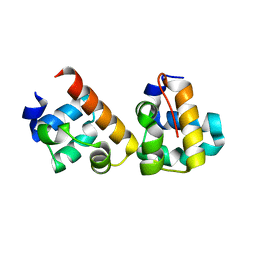 | |
4J19
 
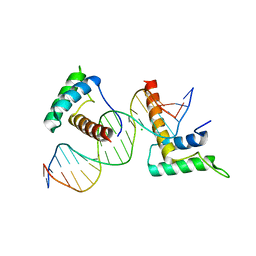 | | Structure of a novel telomere repeat binding protein bound to DNA | | 分子名称: | CHLORIDE ION, DNA (5'-D(*CP*TP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*CP*A)-3'), ... | | 著者 | Kappei, D, Butter, F, Benda, C, Scheibe, M, Draskovic, I, Stevense, M, Lopes Novo, C, Basquin, C, Krastev, D.B, Kittler, R, Jessberger, R, Londono-Vallejo, A.J, Mann, M, Buchholz, F. | | 登録日 | 2013-02-01 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | HOT1 is a mammalian direct telomere repeat-binding protein contributing to telomerase recruitment.
Embo J., 32, 2013
|
|
1ZRZ
 
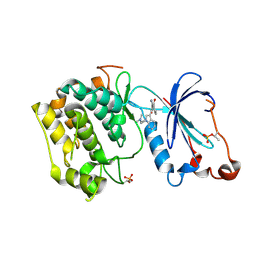 | | Crystal Structure of the Catalytic Domain of Atypical Protein Kinase C-iota | | 分子名称: | 3-{1-[3-(DIMETHYLAMINO)PROPYL]-1H-INDOL-3-YL}-4-(1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, Protein kinase C, iota | | 著者 | Messerschmidt, A, Macieira, S, Velarde, M, Baedeker, M, Benda, C, Jestel, A, Brandstetter, H, Neuefeind, T, Blaesse, M, Structural Proteomics in Europe (SPINE) | | 登録日 | 2005-05-23 | | 公開日 | 2005-09-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of Human Atypical Protein Kinase C-iota Reveals Interaction Mode of Phosphorylation Site in Turn Motif
J.Mol.Biol., 352, 2005
|
|
4N0L
 
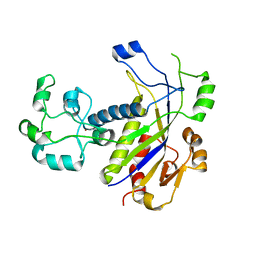 | | Methanopyrus kandleri Csm3 crystal structure | | 分子名称: | Predicted component of a thermophile-specific DNA repair system, contains a RAMP domain, ZINC ION | | 著者 | Hrle, A, Su, A.A, Ebert, J, Benda, C, Conti, E, Randau, L. | | 登録日 | 2013-10-02 | | 公開日 | 2013-11-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Structure and RNA-binding properties of the Type III-A CRISPR-associated protein Csm3.
Rna Biol., 10, 2013
|
|
4JA0
 
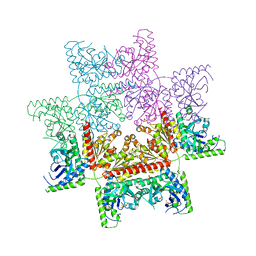 | | Crystal structure of the invertebrate bi-functional purine biosynthesis enzyme PAICS at 2.8 A resolution | | 分子名称: | Phosphoribosylaminoimidazole carboxylase, SULFATE ION | | 著者 | Taschner, M, Basquin, J, Benda, C, Lorentzen, E. | | 登録日 | 2013-02-18 | | 公開日 | 2013-02-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the invertebrate bifunctional purine biosynthesis enzyme PAICS at 2.8 angstrom resolution.
Proteins, 81, 2013
|
|
5OQD
 
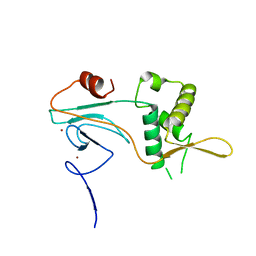 | | PHD2 and winged-helix domain of Polycomblike | | 分子名称: | 1,2-ETHANEDIOL, PHOSPHATE ION, Polycomb protein Pcl, ... | | 著者 | Choi, J, Benda, C, Mueller, J. | | 登録日 | 2017-08-11 | | 公開日 | 2017-11-01 | | 最終更新日 | 2017-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.447 Å) | | 主引用文献 | DNA binding by PHF1 prolongs PRC2 residence time on chromatin and thereby promotes H3K27 methylation.
Nat. Struct. Mol. Biol., 24, 2017
|
|
8PP6
 
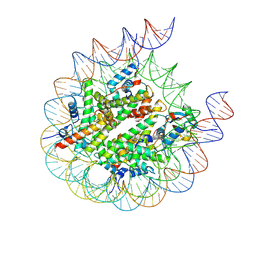 | |
8PP7
 
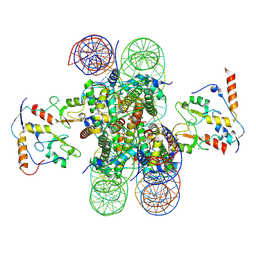 | |
4AJ5
 
 | | Crystal structure of the Ska core complex | | 分子名称: | SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 1, SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 2, SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 3 | | 著者 | Jeyaprakash, A.A, Santamaria, A, Jayachandran, U, Chan, Y.W, Benda, C, Nigg, E.A, Conti, E. | | 登録日 | 2012-02-15 | | 公開日 | 2012-05-23 | | 実験手法 | X-RAY DIFFRACTION (3.32 Å) | | 主引用文献 | Structural and Functional Organization of the Ska Complex, a Key Component of the Kinetochore-Microtubule Interface.
Mol.Cell, 46, 2012
|
|
4CT0
 
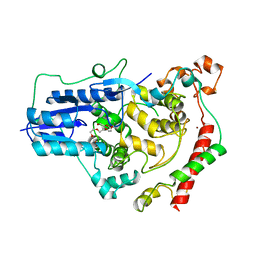 | | Crystal Structure of Mouse Cryptochrome1 in Complex with Period2 | | 分子名称: | CHLORIDE ION, CRYPTOCHROME-1, HEXAETHYLENE GLYCOL, ... | | 著者 | Schmalen, I, Rajan Prabu, J, Benda, C, Wolf, E. | | 登録日 | 2014-03-11 | | 公開日 | 2014-06-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Interaction of Circadian Clock Proteins Cry1 and Per2 is Modulated by Zinc Binding and Disulfide Bond Formation.
Cell(Cambridge,Mass.), 157, 2014
|
|
5LXR
 
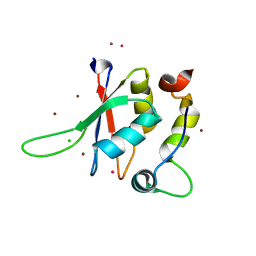 | | Structure of the minimal RBM7 - ZCCHC8 Complex | | 分子名称: | BROMIDE ION, CHLORIDE ION, RNA-binding protein 7, ... | | 著者 | Falk, S, Finogenova, K, Benda, C, Conti, E. | | 登録日 | 2016-09-22 | | 公開日 | 2016-12-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the RBM7-ZCCHC8 core of the NEXT complex reveals connections to splicing factors.
Nat Commun, 7, 2016
|
|
5LXY
 
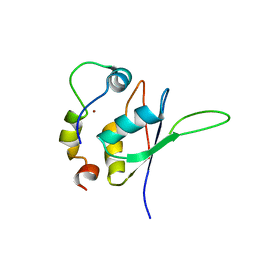 | | Structure of the minimal RBM7 - ZCCHC8 Complex | | 分子名称: | BROMIDE ION, RNA-binding protein 7, Zinc finger CCHC domain-containing protein 8 | | 著者 | Falk, S, Finogenova, K, Benda, C, Conti, E. | | 登録日 | 2016-09-23 | | 公開日 | 2016-12-14 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structure of the RBM7-ZCCHC8 core of the NEXT complex reveals connections to splicing factors.
Nat Commun, 7, 2016
|
|
