5ET8
 
 | | Human muscle fructose-1,6-bisphosphatase in active R-state in complex with fructose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | T-to-R switch of muscle FBPase involves extreme changes of secondary and quaternary structure
To Be Published
|
|
5ET6
 
 | | Human muscle fructose-1,6-bisphosphatase in inactive T-state in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5ET7
 
 | | Human muscle fructose-1,6-bisphosphatase in inactive T-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5ET5
 
 | | Human muscle fructose-1,6-bisphosphatase in active R-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5K56
 
 | | Human muscle fructose-1,6-bisphosphatase in active R-state in complex with fructose-1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
5K54
 
 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in active R-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
5K55
 
 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in active R-state in complex with fructose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
5L0A
 
 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in active R-state in complex with fructose-1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-07-27 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
7R5C
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-29 (G206C, R207S, D210L, S211V) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Isoaspartyl peptidase, ... | | Authors: | Barciszewski, J, Imiolczyk, B, Loch, J.I, Jaskolski, M. | | Deposit date: | 2022-02-10 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6H26
 
 | | Rabbit muscle phosphoglycerate mutase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Phosphoglycerate mutase | | Authors: | Wisniewski, J, Barciszewski, J, Jaskolski, M, Rakus, D. | | Deposit date: | 2018-07-13 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.288 Å) | | Cite: | Rabbit muscle phosphoglycerate mutase
To Be Published
|
|
9HNC
 
 | | Crystal structure of potassium-independent L-asparaginase from Phaseolus vulgaris (PvAIII, PvAspG2) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Loch, J.I, Pierog, I, Imiolczyk, B, Barciszewski, J, Marsolais, F, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-12-10 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | Unique double-helical packing of protein molecules in the crystal of potassium-independent L-asparaginase from common bean.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
3O5W
 
 | | Binding of kinetin in the active site of mistletoe lectin I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Malecki, P.H, Meyer, A, Rypniewski, W, Szymanski, M, Barciszewski, J, Betzel, C. | | Deposit date: | 2010-07-28 | | Release date: | 2011-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding of the plant hormone kinetin in the active site of Mistletoe Lectin I from Viscum album.
Biochim.Biophys.Acta, 1824, 2012
|
|
6ELY
 
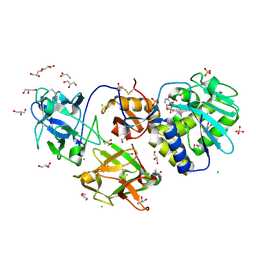 | | Crystal Structure of Mistletoe Lectin I (ML-I) from Viscum album in Complex with 4-N-Furfurylcytosine at 2.84 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-N-Furfurylcytosine, ... | | Authors: | Ahmad, M.S, Rasheed, S, Falke, S, Khaliq, B, Perbandt, M, Choudhary, M.I, Markiewicz, W.T, Barciszewski, J, Betzel, C. | | Deposit date: | 2017-09-30 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal Structure of Mistletoe Lectin I (ML-I) from Viscum album in Complex with 4-N-Furfurylcytosine at 2.85 angstrom Resolution.
Med Chem, 14, 2018
|
|
3D7W
 
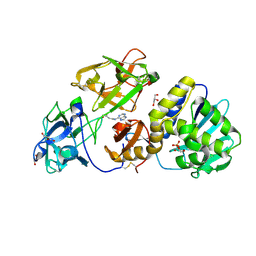 | | Mistletoe Lectin I in Complex with Zeatin | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, (2Z)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Meyer, A, Rypniewski, W, Szymanski, M, Voelter, W, Barciszewski, J, Betzel, C. | | Deposit date: | 2008-05-22 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of mistletoe lectin I from Viscum album in complex with the phytohormone zeatin
Biochim.Biophys.Acta, 1784, 2008
|
|
7O5M
 
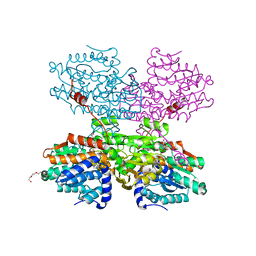 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Na+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Barciszewski, J, Czyrko-Horczak, J, Brzezinski, K. | | Deposit date: | 2021-04-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7O5L
 
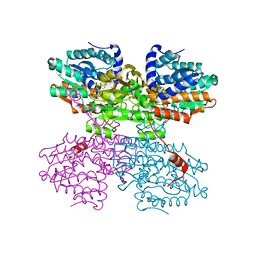 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Barciszewski, J, Czyrko-Horczak, J, Brzezinski, K. | | Deposit date: | 2021-04-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4PPH
 
 | | Crystal structure of conglutin gamma, a unique basic 7S globulin from lupine seeds | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Czubinski, J, Barciszewski, J, Gilski, M, Lampart-Szczapa, E, Jaskolski, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-02-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Structure of gamma-conglutin: insight into the quaternary structure of 7S basic globulins from legumes.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Q0K
 
 | | Crystal Structure of Phytohormone Binding Protein from Medicago truncatula in complex with gibberellic acid (GA3) | | Descriptor: | GIBBERELLIN A3, GLYCEROL, PHYTOHORMONE BINDING PROTEIN MTPHBP | | Authors: | Ciesielska, A, Barciszewski, J, Ruszkowski, M, Jaskolski, M, Sikorski, M. | | Deposit date: | 2014-04-02 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Specific binding of gibberellic acid by Cytokinin-Specific Binding Proteins: a new aspect of plant hormone-binding proteins with the PR-10 fold.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JKX
 
 | | Crystal structure Mistletoe Lectin I from Viscum album in complex with kinetin at 2.35 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Meyer, A, Barciszewski, J, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2013-03-12 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure Mistletoe Lectin I from Viscum album in complex with kinetin at 2.35 A resolution.
To be Published
|
|
4KMG
 
 | | Crystal structure of cytochrome c6B from Synechococcus sp. WH8102 | | Descriptor: | Cytochrome C6 (Soluble cytochrome F) (Cytochrome c553), HEME C, SODIUM ION | | Authors: | Zatwarnicki, P, Krzywda, S, Barciszewski, J, Jaskolski, M, Szczepaniak, A. | | Deposit date: | 2013-05-08 | | Release date: | 2014-03-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cytochrome c6B of Synechococcus sp. WH 8102 - Crystal structure and basic properties of novel c6-like family representative.
Biochem.Biophys.Res.Commun., 443, 2014
|
|
5KY6
 
 | |
7Q2O
 
 | | Beta-lactoglobulin mutant FAW (I56F/L39A/M107W) in complex with desipramine (FAW-DSM#1) | | Descriptor: | 1,2-ETHANEDIOL, 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N-METHYLPROPAN-1-AMINE, Beta-lactoglobulin, ... | | Authors: | Loch, J.I, Barciszewski, J, Lewinski, K. | | Deposit date: | 2021-10-25 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New ligand-binding sites identified in the crystal structures of [beta]-lactoglobulin complexes with desipramine
Iucrj, 9, 2022
|
|
7Q17
 
 | | Beta-lactoglobulin mutant FAW (I56F/L39A/M107W), unliganded form | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactoglobulin | | Authors: | Loch, J.I, Barciszewski, J, Lewinski, K. | | Deposit date: | 2021-10-18 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New ligand-binding sites identified in the crystal structures of [beta]-lactoglobulin complexes with desipramine
Iucrj, 9, 2022
|
|
7Q19
 
 | | Beta-lactoglobulin mutant FAW (I56F/L39A/M107W) in complex with desipramine (FAW-DSM#3) | | Descriptor: | 1,2-ETHANEDIOL, 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N-METHYLPROPAN-1-AMINE, Beta-lactoglobulin, ... | | Authors: | Loch, J.I, Barciszewski, J, Pokrywka, K, Lewinski, K. | | Deposit date: | 2021-10-18 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | New ligand-binding sites identified in the crystal structures of [beta]-lactoglobulin complexes with desipramine
Iucrj, 9, 2022
|
|
7Q2N
 
 | | Beta-lactoglobulin mutant FAF (I56F/L39A/M107F) in complex with desipramine (FAF-DSM) | | Descriptor: | 1,2-ETHANEDIOL, 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N-METHYLPROPAN-1-AMINE, Beta-lactoglobulin, ... | | Authors: | Loch, J.I, Barciszewski, J, Lewinski, K. | | Deposit date: | 2021-10-25 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | New ligand-binding sites identified in the crystal structures of [beta]-lactoglobulin complexes with desipramine
Iucrj, 9, 2022
|
|
