6RM3
 
 | | Evolutionary compaction and adaptation visualized by the structure of the dormant microsporidian ribosome | | 分子名称: | 16S rRNA, 23S rRNA, 5S rRNA, ... | | 著者 | Barandun, J, Hunziker, M, Vossbrinck, C.R, Klinge, S. | | 登録日 | 2019-05-05 | | 公開日 | 2019-07-10 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Evolutionary compaction and adaptation visualized by the structure of the dormant microsporidian ribosome.
Nat Microbiol, 4, 2019
|
|
5WLC
 
 | | The complete structure of the small subunit processome | | 分子名称: | 18S pre-rRNA, 5' ETS, Bms1, ... | | 著者 | Barandun, J, Chaker-Margot, M, Hunziker, M, Klinge, S. | | 登録日 | 2017-07-26 | | 公開日 | 2017-09-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | The complete structure of the small-subunit processome.
Nat. Struct. Mol. Biol., 24, 2017
|
|
4BJR
 
 | | Crystal structure of the complex between Prokaryotic Ubiquitin-like Protein Pup and its Ligase PafA | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PUP--PROTEIN LIGASE, ... | | 著者 | Barandun, J, Delley, C.L, Ban, N, Weber-Ban, E. | | 登録日 | 2013-04-19 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of the Complex between Prokaryotic Ubiquitin-Like Protein Pup and its Ligase Pafa.
J.Am.Chem.Soc., 135, 2013
|
|
2Y1L
 
 | | Caspase-8 in Complex with DARPin-8.4 | | 分子名称: | 1,2-ETHANEDIOL, AC-IETD-CHO, CASPASE-8, ... | | 著者 | Barandun, J, Schroeder, T, Mittl, P.R.E, Grutter, M.G. | | 登録日 | 2010-12-08 | | 公開日 | 2011-12-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Caspase-8 in Complex with Darpin-8.4
To be Published
|
|
2Y0B
 
 | | Caspase-3 in Complex with an Inhibitory DARPin-3.4_S76R | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CASPASE-3, ... | | 著者 | Barandun, J, Schroeder, T, Mittl, P, Grutter, M.G. | | 登録日 | 2010-12-01 | | 公開日 | 2011-12-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Specific Inhibition of Caspase-3 by a Competitive Darpin: Molecular Mimicry between Native and Designed Inhibitors.
Structure, 21, 2013
|
|
2XZT
 
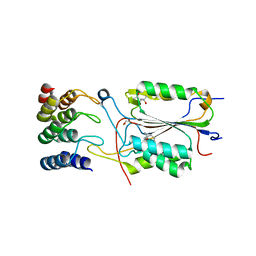 | | Caspase-3 in Complex with DARPin-3.4_I78S | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, CASPASE-3, DARPIN-3.4_I78S | | 著者 | Barandun, J, Schroeder, T, Mittl, P, Grutter, M.G. | | 登録日 | 2010-11-29 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Caspase-3 in Complex with Darpin-3.4_I78S
To be Published
|
|
2XZD
 
 | | Caspase-3 in Complex with an Inhibitory DARPin-3.4 | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CASPASE-3, ... | | 著者 | Barandun, J, Schroeder, T, Mittl, P, Grutter, M.G. | | 登録日 | 2010-11-24 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Specific Inhibition of Caspase-3 by a Competitive Darpin: Molecular Mimicry between Native and Designed Inhibitors.
Structure, 21, 2013
|
|
7OCY
 
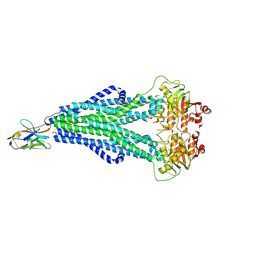 | | Enterococcus faecalis EfrCD in complex with a nanobody | | 分子名称: | ABC transporter ATP-binding protein, Nanobody | | 著者 | Ehrenbolger, K, Hutter, C.A.J, Meier, G, Seeger, M.A, Barandun, J. | | 登録日 | 2021-04-28 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (4.25 Å) | | 主引用文献 | Deep mutational scan of a drug efflux pump reveals its structure-function landscape.
Nat.Chem.Biol., 19, 2023
|
|
7P3R
 
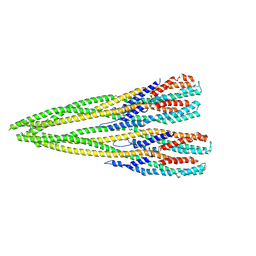 | | Helical structure of the toxin MakA from Vibrio cholera | | 分子名称: | MakA tetramer | | 著者 | Berg, A, Nadeem, A, Uhlin, B.E, Wai, S.N, Barandun, J. | | 登録日 | 2021-07-08 | | 公開日 | 2022-02-23 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | Protein-lipid interaction at low pH induces oligomerization of the MakA cytotoxin from Vibrio cholerae .
Elife, 11, 2022
|
|
8ADN
 
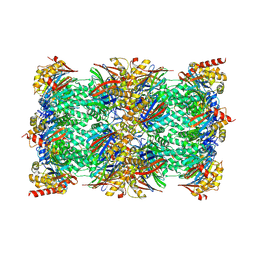 | | Vairimorpha necatrix 20S proteasome from spores | | 分子名称: | Proteasome Inhibitor 31-Like, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | 著者 | Jespersen, N, Ehrenbolger, K, Winiger, R, Svedberg, D, Vossbrinck, C.R, Barandun, J. | | 登録日 | 2022-07-08 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Structure of the reduced microsporidian proteasome bound by PI31-like peptides in dormant spores.
Nat Commun, 13, 2022
|
|
6ZU5
 
 | | Structure of the Paranosema locustae ribosome in complex with Lso2 | | 分子名称: | 18S rRNA, 25S rRNA, 5S rRNA, ... | | 著者 | Ehrenbolger, K, Jespersen, N, Sharma, H, Sokolova, Y.Y, Tokarev, Y.S, Vossbrinck, C.R, Barandun, J. | | 登録日 | 2020-07-21 | | 公開日 | 2020-11-04 | | 最終更新日 | 2020-11-11 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Differences in structure and hibernation mechanism highlight diversification of the microsporidian ribosome.
Plos Biol., 18, 2020
|
|
6ND4
 
 | | Conformational switches control early maturation of the eukaryotic small ribosomal subunit | | 分子名称: | 18S rRNA 5' domain start, 5'ETS rRNA, Bud21, ... | | 著者 | Hunziker, M, Barandun, J, Klinge, S. | | 登録日 | 2018-12-13 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Conformational switches control early maturation of the eukaryotic small ribosomal subunit.
Elife, 8, 2019
|
|
6CB1
 
 | | Yeast nucleolar pre-60S ribosomal subunit (state 3) | | 分子名称: | 35S pre-ribosomal RNA miscRNA, 5.8S rRNA, 60S ribosomal protein L13-A, ... | | 著者 | Sanghai, Z.A, Miller, L, Barandun, J, Hunziker, M, Chaker-Margot, M, Klinge, S. | | 登録日 | 2018-02-01 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Modular assembly of the nucleolar pre-60S ribosomal subunit.
Nature, 556, 2018
|
|
6C0F
 
 | | Yeast nucleolar pre-60S ribosomal subunit (state 2) | | 分子名称: | 5.8S rRNA, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | 著者 | Sanghai, Z.A, Miller, L, Barandun, J, Hunziker, M, Chaker-Margot, M, Klinge, S. | | 登録日 | 2017-12-29 | | 公開日 | 2018-03-14 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Modular assembly of the nucleolar pre-60S ribosomal subunit.
Nature, 556, 2018
|
|
5TZS
 
 | | Architecture of the yeast small subunit processome | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 3' domain-associated, ... | | 著者 | Chaker-Margot, M, Barandun, J, Hunziker, M, Klinge, S. | | 登録日 | 2016-11-22 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Architecture of the yeast small subunit processome.
Science, 355, 2017
|
|
4B0S
 
 | | Structure of the Deamidase-Depupylase Dop of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DEAMIDASE-DEPUPYLASE DOP, MAGNESIUM ION | | 著者 | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | 登録日 | 2012-07-04 | | 公開日 | 2012-09-12 | | 最終更新日 | 2020-04-01 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structures of Pup Ligase Pafa and Depupylase Dop from the Prokaryotic Ubiquitin-Like Modification Pathway.
Nat.Commun., 3, 2012
|
|
4B0R
 
 | | Structure of the Deamidase-Depupylase Dop of the Prokaryotic Ubiquitin-like Modification Pathway | | 分子名称: | DEAMIDASE-DEPUPYLASE DOP | | 著者 | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | 登録日 | 2012-07-04 | | 公開日 | 2012-09-12 | | 最終更新日 | 2019-02-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structures of Pup ligase PafA and depupylase Dop from the prokaryotic ubiquitin-like modification pathway.
Nat Commun, 3, 2012
|
|
4B0T
 
 | | Structure of the Pup Ligase PafA of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PUP--PROTEIN LIGASE | | 著者 | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | 登録日 | 2012-07-04 | | 公開日 | 2012-09-12 | | 実験手法 | X-RAY DIFFRACTION (2.159 Å) | | 主引用文献 | Structures of Pup Ligase Pafa and Depupylase Dop from the Prokaryotic Ubiquitin-Like Modification Pathway.
Nat.Commun., 3, 2012
|
|
