2MZK
 
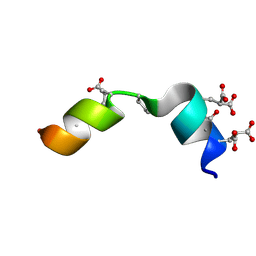 | | The solution structure of the Magnesium-bound Conantokin RLB | | Descriptor: | Conantokin-R1-B | | Authors: | Kunda, S, Yuan, Y, Balsara, R.D, Zajicek, J, Castellino, F.J. | | Deposit date: | 2015-02-14 | | Release date: | 2015-06-17 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Hydroxyproline-induced Helical Disruption in Conantokin Rl-B Affects Subunit-selective Antagonistic Activities toward Ion Channels of N-Methyl-d-aspartate Receptors.
J.Biol.Chem., 290, 2015
|
|
2MYZ
 
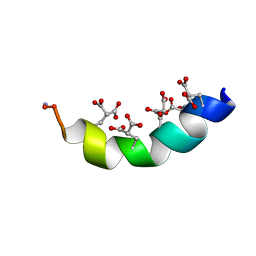 | | The Solution Structure of the Magnesium-bound Conantokin-R1B Mutant | | Descriptor: | Conantokin-R1-B | | Authors: | Kunda, S, Yuan, Y, Balsara, R.D, Zajicek, J, Castellino, F.J. | | Deposit date: | 2015-02-04 | | Release date: | 2015-06-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Hydroxyproline-induced Helical Disruption in Conantokin Rl-B Affects Subunit-selective Antagonistic Activities toward Ion Channels of N-Methyl-d-aspartate Receptors.
J.Biol.Chem., 290, 2015
|
|
2MZM
 
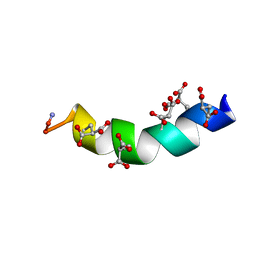 | | The Solution Structure of the Magnesium-bound Conantokin-G | | Descriptor: | Conantokin-R1-B | | Authors: | Kunda, S, Yuan, Y, Balsara, R.D, Zajicek, J, Castellino, F.J. | | Deposit date: | 2015-02-14 | | Release date: | 2015-06-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Hydroxyproline-induced Helical Disruption in Conantokin Rl-B Affects Subunit-selective Antagonistic Activities toward Ion Channels of N-Methyl-d-aspartate Receptors.
J.Biol.Chem., 290, 2015
|
|
2MZL
 
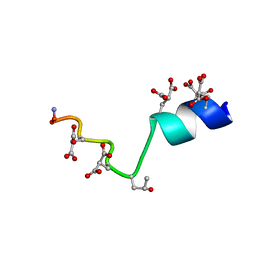 | | The Solution Structure of the Magnesium-bound Conantokin-G Mutant | | Descriptor: | Conantokin-R1-B | | Authors: | Kunda, S, Yuan, Y, Balsara, R.D, Zajicek, J, Castellino, F.J. | | Deposit date: | 2015-02-14 | | Release date: | 2015-06-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Hydroxyproline-induced Helical Disruption in Conantokin Rl-B Affects Subunit-selective Antagonistic Activities toward Ion Channels of N-Methyl-d-aspartate Receptors.
J.Biol.Chem., 290, 2015
|
|
5TBQ
 
 | |
5TBR
 
 | |
5TBG
 
 | |
6O8S
 
 | | Syn-safencin 56 | | Descriptor: | Circular bacteriocin, circularin A/uberolysin family | | Authors: | Fields, F.R, Lee, S.W. | | Deposit date: | 2019-03-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synthetic Antimicrobial Peptide Tuning Permits Membrane Disruption and Interpeptide Synergy.
Acs Pharmacol Transl Sci, 3, 2020
|
|
6O8T
 
 | | Syn-safencin 96 | | Descriptor: | Circular bacteriocin, circularin A/uberolysin family | | Authors: | Fields, F.R, Lee, S.W. | | Deposit date: | 2019-03-11 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Synthetic Antimicrobial Peptide Tuning Permits Membrane Disruption and Interpeptide Synergy.
Acs Pharmacol Transl Sci, 3, 2020
|
|
6O8R
 
 | | Syn-safencin 24 | | Descriptor: | Circular bacteriocin, circularin A/uberolysin family | | Authors: | Fields, F.R, Lee, S.W. | | Deposit date: | 2019-03-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synthetic Antimicrobial Peptide Tuning Permits Membrane Disruption and Interpeptide Synergy.
Acs Pharmacol Transl Sci, 3, 2020
|
|
6O8J
 
 | | Syn-safencin | | Descriptor: | Circular bacteriocin, circularin A/uberolysin family | | Authors: | Fields, F.R, Lee, S.W. | | Deposit date: | 2019-03-11 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synthetic Antimicrobial Peptide Tuning Permits Membrane Disruption and Interpeptide Synergy.
Acs Pharmacol Transl Sci, 3, 2020
|
|
6O8P
 
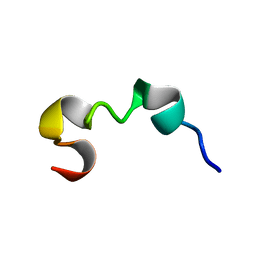 | | Syn-safencin 8 | | Descriptor: | Circular bacteriocin, circularin A/uberolysin family | | Authors: | Fields, F.R, Lee, S.W. | | Deposit date: | 2019-03-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synthetic Antimicrobial Peptide Tuning Permits Membrane Disruption and Interpeptide Synergy.
Acs Pharmacol Transl Sci, 3, 2020
|
|
6BZL
 
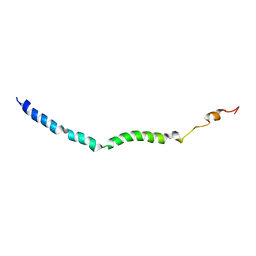 | | Solution structure of VEK75 | | Descriptor: | M protein | | Authors: | Qiu, C, Yuan, Y, Castellino, F.J. | | Deposit date: | 2017-12-24 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Contributions of different modules of the plasminogen-binding Streptococcus pyogenes M-protein that mediate its functional dimerization.
J.Struct.Biol., 204, 2018
|
|
6BZJ
 
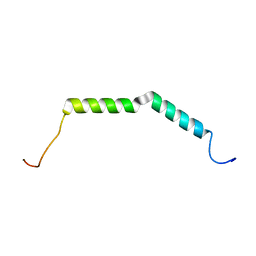 | | Solution structure of AGL55 | | Descriptor: | M protein | | Authors: | Qiu, C, Yuan, Y, Castellino, F.J. | | Deposit date: | 2017-12-24 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Contributions of different modules of the plasminogen-binding Streptococcus pyogenes M-protein that mediate its functional dimerization.
J.Struct.Biol., 204, 2018
|
|
6BZK
 
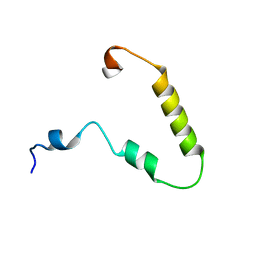 | | Solution structure of KTI55 | | Descriptor: | M protein | | Authors: | Castellino, F.J, Qiu, C, Yuan, Y. | | Deposit date: | 2017-12-24 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Contributions of different modules of the plasminogen-binding Streptococcus pyogenes M-protein that mediate its functional dimerization.
J.Struct.Biol., 204, 2018
|
|
