1ZHN
 
 | | Crystal Structure of mouse CD1d bound to the self ligand phosphatidylcholine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(DODECANOYLOXY)METHYL]-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-3,5,8-TRIOXA-4-PHOSPHADOTRIACONTAN-1-AMINIUM 4-OXIDE, CD1d1 antigen, ... | | Authors: | Giabbai, B, Sidobre, S, Crispin, M.M.D, Sanchez Ruiz, Y, Bachi, A, Kronenberg, M, Wilson, I.A, Degano, M. | | Deposit date: | 2005-04-26 | | Release date: | 2005-07-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: a molecular basis for NKT cell activation
J.Immunol., 175, 2005
|
|
4CFM
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 6-(cyclohexylmethoxy)-8-(2-methylphenyl)-9H-purin-2-amine, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-18 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
4CFW
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 3-[2-amino-6-(cyclohexylmethoxy)-7H-purin-8-yl]-2-methylbenzenesulfonamide, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
4CFU
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 3-[2-azanyl-6-(cyclohexylmethoxy)-7H-purin-8-yl]-2-methyl-benzoic acid, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2, ... | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
4CFV
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 3-[2-amino-6-(cyclohexylmethoxy)-7H-purin-8-yl]-2-methylphenol, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2, ... | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2014-12-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
4CFX
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 3-[2-amino-6-(cyclohexylmethoxy)-7H-purin-8-yl]benzenesulfonamide, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2014-12-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
4CFN
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6-(cyclohexylmethoxy)-8-(trifluoromethyl)-9H-purin-2-amine, CYCLIN-A2, ... | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
8H5C
 
 | | Structure of SARS-CoV-2 Omicron BA.2.75 RBD in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Bai, B, Liu, K.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-10-12 | | Release date: | 2023-07-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7OY5
 
 | | Crystal structure of GSK3Beta in complex with ARN25068 | | Descriptor: | CHLORIDE ION, Glycogen synthase kinase-3 beta, ~{N}4-(3-cyclopropyl-1~{H}-pyrazol-5-yl)-~{N}2-(phenylmethyl)thieno[3,2-d]pyrimidine-2,4-diamine | | Authors: | Tripathi, S.K, Balboni, B, Demuro, S, DiMartino, R, Giabbai, B, Storici, P, Ortega, J, Girotto, S, Cavalli, A. | | Deposit date: | 2021-06-23 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | ARN25068, a versatile starting point towards triple GSK-3 beta /FYN/DYRK1A inhibitors to tackle tau-related neurological disorders.
Eur.J.Med.Chem., 229, 2022
|
|
3MKM
 
 | | Crystal structure of the E. coli pyrimidine nucleoside hydrolase YeiK (Apo-form) | | Descriptor: | CALCIUM ION, Putative uncharacterized protein YeiK | | Authors: | Garau, G, Fornili, A, Giabbai, B, Degano, M. | | Deposit date: | 2010-04-15 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Energy Landscapes Associated with Macromolecular Conformational Changes from Endpoint Structures
J.Am.Chem.Soc., 132, 2010
|
|
3MKN
 
 | | Crystal structure of the E. coli pyrimidine nucleosidase YeiK bound to a competitive inhibitor | | Descriptor: | (2S,3S,4R,5R)-2-(3,4-diaminophenyl)-5-(hydroxymethyl)pyrrolidine-3,4-diol, CALCIUM ION, Putative uncharacterized protein YeiK | | Authors: | Garau, G, Fornili, A, Giabbai, B, Degano, M. | | Deposit date: | 2010-04-15 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Energy Landscapes Associated with Macromolecular Conformational Changes from Endpoint Structures
J.Am.Chem.Soc., 132, 2010
|
|
6HK4
 
 | | Crystal structure of GSK-3B in complex with pyrazine inhibitor C22 | | Descriptor: | 3-azanyl-6-(4-morpholin-4-ylsulfonylphenyl)-~{N}-pyridin-3-yl-pyrazine-2-carboxamide, DIMETHYL SULFOXIDE, GLY-SER-HIS-GLY-HIS-HIS-HIS-HIS-HIS, ... | | Authors: | Piretti, V, Giabbai, B, Demitri, N, Di Martino, R, Tripathi, S.K, Gobbo, D, Decherchi, S, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2018-09-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Investigating Drug-Target Residence Time in Kinases through Enhanced Sampling Simulations.
J Chem Theory Comput, 15, 2019
|
|
6HK3
 
 | | Crystal structure of GSK-3B in complex with pyrazine inhibitor C44 | | Descriptor: | 3-azanyl-~{N}-(2-methoxyphenyl)-6-[4-(4-methylpiperazin-1-yl)sulfonylphenyl]pyrazine-2-carboxamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Piretti, V, Giabbai, B, Demitri, N, Di Martino, R, Tripathi, S.K, Gobbo, D, Decherchi, S, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2018-09-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Investigating Drug-Target Residence Time in Kinases through Enhanced Sampling Simulations.
J Chem Theory Comput, 15, 2019
|
|
6HK7
 
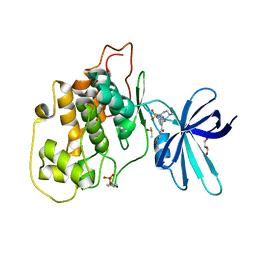 | | Crystal structure of GSK-3B in complex with pyrazine inhibitor C50 | | Descriptor: | 3-azanyl-~{N}-(2-methoxyethyl)-6-[4-(4-methylpiperazin-1-yl)sulfonylphenyl]pyrazine-2-carboxamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Piretti, V, Giabbai, B, Demitri, N, Di Martino, R, Tripathi, S.K, Gobbo, D, Decherchi, S, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2018-09-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Investigating Drug-Target Residence Time in Kinases through Enhanced Sampling Simulations.
J Chem Theory Comput, 15, 2019
|
|
7NBY
 
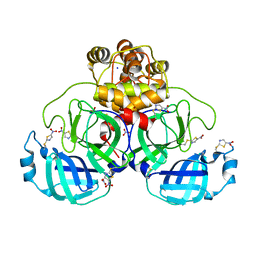 | | Crystal structure of SU3327 (halicin) covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | 5-nitro-1,3-thiazole, CHLORIDE ION, Main Protease, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of SU3327 (halicin) covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2.
To Be Published
|
|
7NG6
 
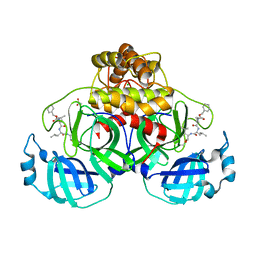 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P1 in absence of DTT. | | Descriptor: | 3C-like proteinase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7NG3
 
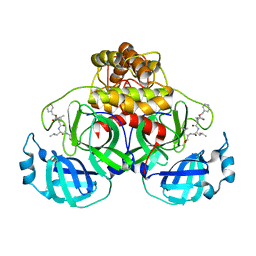 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P1. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7NF5
 
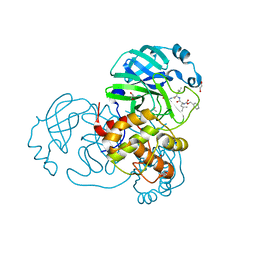 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup C2. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-05 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7B3E
 
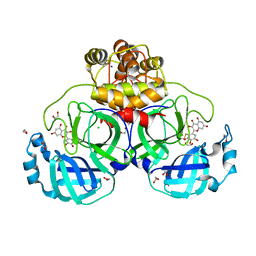 | | Crystal structure of myricetin covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2020-11-30 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Identification of Inhibitors of SARS-CoV-2 3CL-Pro Enzymatic Activity Using a Small Molecule in Vitro Repurposing Screen.
Acs Pharmacol Transl Sci, 4, 2021
|
|
5NEV
 
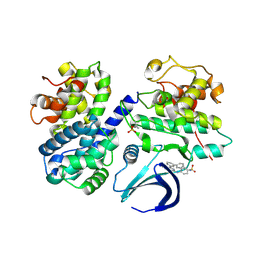 | | CDK2/Cyclin A in complex with compound 73 | | Descriptor: | 4-[[6-(3-phenylphenyl)-7~{H}-purin-2-yl]amino]benzenesulfonamide, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Coxon, C.R, Anscombe, E, Harnor, S.J, Martin, M.P, Carbain, B, Hardcastle, I.R, Harlow, L.K, Korolchuk, S, Matheson, C.J, Noble, M.E.M, Newell, D.R, Turner, D, Sivaprakasam, M, Wang, L.Z, Wong, C, Golding, B.T, Griffin, R.J, Cano, G. | | Deposit date: | 2017-03-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Cyclin-Dependent Kinase (CDK) Inhibitors: Structure-Activity Relationships and Insights into the CDK-2 Selectivity of 6-Substituted 2-Arylaminopurines.
J. Med. Chem., 60, 2017
|
|
6BR5
 
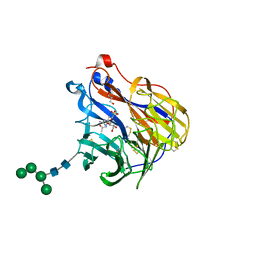 | | N2 neuraminidase in complex with a novel antiviral compound | | Descriptor: | (1R)-4-acetamido-1,5-anhydro-3-carbamimidamido-2,3,4-trideoxy-1-sulfo-D-glycero-D-galacto-octitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, L.H, Ve, T, Pascolutti, M, Hadhazi, A, Bailly, B, Thomson, R.J, Gao, G.F, von Itzstein, M. | | Deposit date: | 2017-11-30 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.0379076 Å) | | Cite: | A Sulfonozanamivir Analogue Has Potent Anti-influenza Virus Activity.
ChemMedChem, 13, 2018
|
|
7BFB
 
 | | Crystal structure of ebselen covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | 1,2-ETHANEDIOL, Main Protease, N-phenyl-2-selanylbenzamide, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-01-02 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of ebselen covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2.
To Be Published
|
|
7BE7
 
 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7BGP
 
 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in absence of DTT. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-01-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7BB2
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.6A resolution (spacegroup P2(1)2(1)2(1)) | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2020-12-16 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
