6L9U
 
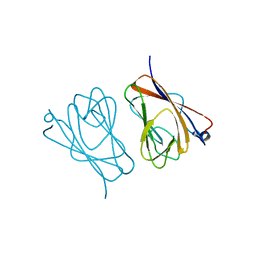 | | Crystal structure of mouse TIFA | | Descriptor: | TRAF-interacting protein with FHA domain-containing protein A | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2019-11-11 | | Release date: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural analysis of TIFA: Insight into TIFA-dependent signal transduction in innate immunity.
Sci Rep, 10, 2020
|
|
7WCE
 
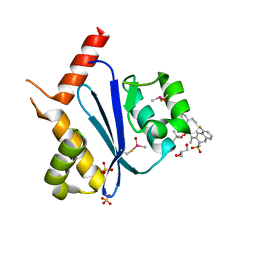 | | Crystal structure of HIV-1 integrase catalytic core domain in complex with (2S)-2-(tert-Butoxy)-2-(10-fluoro-2-(2-hydroxy-4-methylphenyl)-1,4-dimethyl-5-(methylsulfonyl)-5,6-dihydrophenanthridin-3-yl)acetic acid | | Descriptor: | (2S)-2-[10-fluoranyl-1,4-dimethyl-2-(4-methyl-2-oxidanyl-phenyl)-5-methylsulfonyl-6H-phenanthridin-3-yl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, GLYCEROL, Integrase catalytic, ... | | Authors: | Taoda, Y, Sekiguchi, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of tricyclic HIV-1 integrase-LEDGF/p75 allosteric inhibitors by intramolecular direct arylation reaction.
Bioorg.Med.Chem.Lett., 64, 2022
|
|
3AU3
 
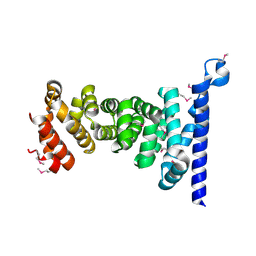 | | Crystal structure of armadillo repeat domain of APC | | Descriptor: | Adenomatous polyposis coli protein | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-01-28 | | Release date: | 2011-11-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the armadillo repeat domain of adenomatous polyposis coli and its complex with the tyrosine-rich domain of sam68
Structure, 19, 2011
|
|
7D83
 
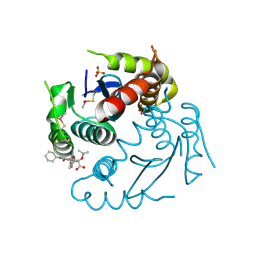 | | Crystal structure of HIV-1 integrase catalytic core domain in complex with 2-(tert-butoxy)-2-(2-(3-cyclohexylureido)-3,6-dimethyl-5-(5-methylchroman-6-yl)pyridin-4-yl)acetic acid | | Descriptor: | (2S)-2-[2-(cyclohexylcarbamoylamino)-3,6-dimethyl-5-(5-methyl-3,4-dihydro-2H-chromen-6-yl)pyridin-4-yl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, Integrase, SULFATE ION | | Authors: | Sugiyama, S, Sekiguchi, Y. | | Deposit date: | 2020-10-07 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of novel HIV-1 integrase-LEDGF/p75 allosteric inhibitors based on a pyridine scaffold forming an intramolecular hydrogen bond.
Bioorg.Med.Chem.Lett., 33, 2020
|
|
7DQE
 
 | | Crystal structure of the ADP-bound mutant A(S23C)3B(N64C)3 complex from enterococcus hirae V-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Maruyama, S, Nakamoto, K, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, T. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | The Combination of High-Speed AFM and X-ray Crystallography Reveals Rotary Catalytic Mechanism of Shaftless V1-ATPase
To Be Published
|
|
7DQD
 
 | | Crystal structure of the AMP-PNP-bound mutant A(S23C)3B(N64C)3 complex from enterococcus hirae V-ATPase | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, T. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.383 Å) | | Cite: | The combination of high-speed AFM and X-ray crystallography reveals rotary catalysis
To Be Published
|
|
7DQC
 
 | | Crystal structure of nucleotide-free mutant A(S23C)3B(N64C)3 complex from Enterococcus hirae V-ATPase | | Descriptor: | GLYCEROL, V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | The combination of high-speed AFM and X-ray crystallography reveals rotary catalytic mechanism of shaftless V1-ATPase
To Be Published
|
|
5B71
 
 | | Crystal structure of complement C5 in complex with SKY59 | | Descriptor: | Complement C5 beta chain, SKY59 Fab heavy chain, SKY59 Fab light chain | | Authors: | Irie, M, Shimizu, Y, Sampei, Z, Fukuzawa, T. | | Deposit date: | 2016-06-03 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Long lasting neutralization of C5 by SKY59, a novel recycling antibody, is a potential therapy for complement-mediated diseases.
Sci Rep, 7, 2017
|
|
