1HRE
 
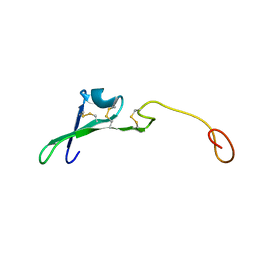 | | SOLUTION STRUCTURE OF THE EPIDERMAL GROWTH FACTOR-LIKE DOMAIN OF HEREGULIN-ALPHA, A LIGAND FOR P180ERB4 | | Descriptor: | HEREGULIN ALPHA | | Authors: | Nagata, K, Kohda, D, Hatanaka, H, Ichikawa, S, Inagaki, F. | | Deposit date: | 1994-07-21 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the epidermal growth factor-like domain of heregulin-alpha, a ligand for p180erbB-4.
EMBO J., 13, 1994
|
|
1HRF
 
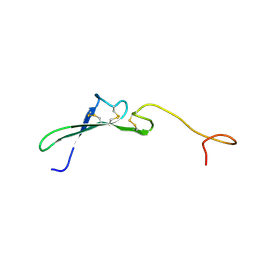 | | SOLUTION STRUCTURE OF THE EPIDERMAL GROWTH FACTOR-LIKE DOMAIN OF HEREGULIN-ALPHA, A LIGAND FOR P180ERB4 | | Descriptor: | HEREGULIN ALPHA | | Authors: | Nagata, K, Kohda, D, Hatanaka, H, Ichikawa, S, Inagaki, F. | | Deposit date: | 1994-07-21 | | Release date: | 1994-10-15 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the epidermal growth factor-like domain of heregulin-alpha, a ligand for p180erbB-4.
EMBO J., 13, 1994
|
|
4L3O
 
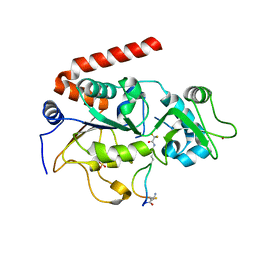 | | Crystal Structure of SIRT2 in complex with the macrocyclic peptide S2iL5 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Yamagata, K, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2013-06-06 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.518 Å) | | Cite: | Structural Basis for Potent Inhibition of SIRT2 Deacetylase by a Macrocyclic Peptide Inducing Dynamic Structural Change
Structure, 22, 2013
|
|
4HE7
 
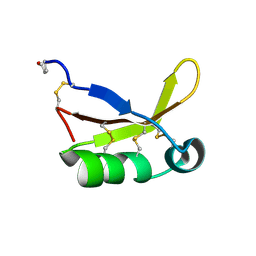 | | Crystal Structure of Brazzein | | Descriptor: | Defensin-like protein, SODIUM ION | | Authors: | Nagata, K, Hongo, N, Kameda, Y, Yamamura, A, Sasaki, H, Lee, W.C, Ishikawa, K, Suzuki, E, Tanokura, M. | | Deposit date: | 2012-10-03 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of brazzein, a sweet-tasting protein from the wild African plant Pentadiplandra brazzeana
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1ZOV
 
 | | Crystal Structure of Monomeric Sarcosine Oxidase from Bacillus sp. NS-129 | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric sarcosine oxidase | | Authors: | Nagata, K, Sasaki, H, Ohtsuka, J, Hua, M, Okai, M, Kubota, K, Kamo, M, Ito, K, Ichikawa, T, Koyama, Y, Tanokura, M. | | Deposit date: | 2005-05-14 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of monomeric sarcosine oxidase from Bacillus sp. NS-129 reveals multiple conformations at the active-site loop
PROC.JPN.ACAD.,SER.B, 81, 2005
|
|
1V46
 
 | |
1BOM
 
 | | THREE-DIMENSIONAL STRUCTURE OF BOMBYXIN-II, AN INSULIN-RELATED BRAIN-SECRETORY PEPTIDE OF THE SILKMOTH BOMBYX MORI: COMPARISON WITH INSULIN AND RELAXIN | | Descriptor: | BOMBYXIN-II,BOMBYXIN A-2, BOMBYXIN-II,BOMBYXIN A-6 | | Authors: | Nagata, K, Hatanaka, H, Kohda, D, Inagaki, F. | | Deposit date: | 1994-07-21 | | Release date: | 1994-11-01 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of bombyxin-II an insulin-like peptide of the silkmoth Bombyx mori: structural comparison with insulin and relaxin.
J.Mol.Biol., 253, 1995
|
|
1BON
 
 | | THREE-DIMENSIONAL STRUCTURE OF BOMBYXIN-II, AN INSULIN-RELATED BRAIN-SECRETORY PEPTIDE OF THE SILKMOTH BOMBYX MORI: COMPARISON WITH INSULIN AND RELAXIN | | Descriptor: | BOMBYXIN-II,BOMBYXIN A-2, BOMBYXIN-II,BOMBYXIN A-6 | | Authors: | Nagata, K, Hatanaka, H, Kohda, D, Inagaki, F, Structural Proteomics in Europe (SPINE) | | Deposit date: | 1994-07-21 | | Release date: | 1995-01-26 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of bombyxin-II an insulin-like peptide of the silkmoth Bombyx mori: structural comparison with insulin and relaxin.
J.Mol.Biol., 253, 1995
|
|
6KZA
 
 | | Crystal structure of the complex of the interaction domains of E. coli DnaB helicase and DnaC helicase loader | | Descriptor: | DNA replication protein DnaC, Replicative DNA helicase | | Authors: | Nagata, K, Okada, A, Ohtsuka, J, Ohkuri, T, Akama, Y, Sakiyama, Y, Miyazaki, E, Horita, S, Katayama, T, Ueda, T, Tanokura, M. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the complex of the interaction domains of Escherichia coli DnaB helicase and DnaC helicase loader: structural basis implying a distortion-accumulation mechanism for the DnaB ring opening caused by DnaC binding.
J.Biochem., 167, 2020
|
|
1Y49
 
 | |
1EQK
 
 | | SOLUTION STRUCTURE OF ORYZACYSTATIN-I, A CYSTEINE PROTEINASE INHIBITOR OF THE RICE, ORYZA SATIVA L. JAPONICA | | Descriptor: | ORYZACYSTATIN-I | | Authors: | Nagata, K, Kudo, N, Abe, K, Arai, S, Tanokura, M. | | Deposit date: | 2000-04-05 | | Release date: | 2001-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of oryzacystatin-I, a cysteine proteinase inhibitor of the rice, Oryza sativa L. japonica.
Biochemistry, 39, 2000
|
|
2YQY
 
 | | Crystal structure of TT2238, a four-helix bundle protein | | Descriptor: | Hypothetical protein TTHA0303 | | Authors: | Nagata, K, Ohtsuka, J, Iino, H, Ebihara, A, Yokoyama, S, Kuramitsu, S, Tanokura, M. | | Deposit date: | 2007-03-31 | | Release date: | 2008-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of TTHA0303 (TT2238), a four-helix bundle protein with an exposed histidine triad from Thermus thermophilus HB8 at 2.0 A
Proteins, 70, 2008
|
|
1WRB
 
 | | Crystal structure of the N-terminal RecA-like domain of DjVLGB, a pranarian Vasa-like RNA helicase | | Descriptor: | DjVLGB, SULFATE ION | | Authors: | Kurimoto, K, Muto, Y, Obayashi, N, Terada, T, Shirouzu, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Kigawa, T, Okumura, H, Tanaka, A, Shibata, N, Kashikawa, M, Agata, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-10-14 | | Release date: | 2005-04-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the N-terminal RecA-like domain of a DEAD-box RNA helicase, the Dugesia japonica vasa-like gene B protein
J.Struct.Biol., 150, 2005
|
|
5WVU
 
 | | Crystal structure of carboxypeptidase from Thermus thermophilus | | Descriptor: | GLYCEROL, Thermostable carboxypeptidase 1, ZINC ION | | Authors: | Okai, M, Nagata, K, Tanokura, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the transition between the open and closed conformations of Thermus thermophilus carboxypeptidase.
Biochem. Biophys. Res. Commun., 484, 2017
|
|
8J4H
 
 | | X-ray structure of a ferric ion-binding protein A (FbpA) from Vibrio metschnikovii in complex with Danshensu (DSS) | | Descriptor: | (2~{R})-3-[3,4-bis(oxidanyl)phenyl]-2-oxidanyl-propanoic acid, Ferric iron ABC transporter iron-binding protein | | Authors: | Lu, P, Jiang, J, Nagata, K. | | Deposit date: | 2023-04-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Molecular mechanism of Fe 3+ binding inhibition to Vibrio metschnikovii ferric ion-binding protein, FbpA, by rosmarinic acid and its hydrolysate, danshensu.
Protein Sci., 33, 2024
|
|
8J4J
 
 | | X-ray structure of a ferric ion-binding protein A (FbpA) from Vibrio metschnikovii in complex with ferric ion | | Descriptor: | CARBONATE ION, FE (III) ION, Ferric iron ABC transporter iron-binding protein | | Authors: | Lu, P, Jiang, J, Nagata, K. | | Deposit date: | 2023-04-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular mechanism of Fe 3+ binding inhibition to Vibrio metschnikovii ferric ion-binding protein, FbpA, by rosmarinic acid and its hydrolysate, danshensu.
Protein Sci., 33, 2024
|
|
8GNN
 
 | | Crystal structure of the human RAD9-RAD1-HUS1-RAD17 complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Cell cycle checkpoint protein RAD17, ... | | Authors: | Hara, K, Nagata, K, Iida, N, Hashimoto, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | The 9-1-1 DNA clamp subunit RAD1 forms specific interactions with clamp loader RAD17, revealing functional implications for binding-protein RHINO.
J.Biol.Chem., 299, 2023
|
|
3GQB
 
 | | Crystal Structure of the A3B3 complex from V-ATPase | | Descriptor: | V-type ATP synthase alpha chain, V-type ATP synthase beta chain | | Authors: | Meher, M, Akimoto, S, Iwata, M, Nagata, K, Hori, Y, Yoshida, M, Yokoyama, S, Iwata, S, Yokoyama, K. | | Deposit date: | 2009-03-24 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of A(3)B(3) complex of V-ATPase from Thermus thermophilus.
Embo J., 28, 2009
|
|
3LNQ
 
 | | Structure of Aristaless homeodomain in complex with DNA | | Descriptor: | 5'-D(*CP*CP*CP*TP*AP*AP*TP*TP*AP*AP*AP*CP*CP*C)-3', 5'-D(*GP*GP*GP*TP*TP*TP*AP*AP*TP*TP*AP*GP*GP*G)-3', ACETATE ION, ... | | Authors: | Takamura, Y, Miyazono, K, Nagata, K, Saigo, K, Kojima, T, Tanokura, M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cooperative DNA-binding and sequence-recognition mechanism of aristaless and clawless
Embo J., 29, 2010
|
|
8J8D
 
 | |
8J8T
 
 | |
3LQB
 
 | | Crystal structure of the hatching enzyme ZHE1 from the zebrafish Danio rerio | | Descriptor: | 1,2-ETHANEDIOL, LOC792177 protein, SULFATE ION, ... | | Authors: | Tanokura, M, Okada, A, Nagata, K, Yasumasu, S, Ohtsuka, J, Iuchi, I. | | Deposit date: | 2010-02-08 | | Release date: | 2010-09-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of zebrafish hatching enzyme 1 from the zebrafish Danio rerio
J.Mol.Biol., 402, 2010
|
|
8K4R
 
 | | Structure of VinM-VinL complex | | Descriptor: | Acyl-carrier-protein, Non-ribosomal peptide synthetase, SODIUM ION, ... | | Authors: | Miyanaga, A, Nagata, K, Nakajima, J, Chisuga, T, Kudo, F, Eguchi, T. | | Deposit date: | 2023-07-20 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Amide-Forming Adenylation Enzyme VinM in Vicenistatin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
4TMC
 
 | | CRYSTAL STRUCTURE of OLD YELLOW ENZYME from CANDIDA MACEDONIENSIS AKU4588 COMPLEXED with P-HYDROXYBENZALDEHYDE | | Descriptor: | FLAVIN MONONUCLEOTIDE, Old yellow enzyme, P-HYDROXYBENZALDEHYDE | | Authors: | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2014-05-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Engineered Old Yellow Enzyme that Enables Efficient Synthesis of (4R,6R)-Actinol in a One-Pot Reduction System
Chembiochem, 16, 2015
|
|
1X0F
 
 | | Complex structure of the C-terminal RNA-binding domain of hnRNP D(AUF1) with telomeric DNA | | Descriptor: | 5'-D(P*TP*AP*GP*G)-3', Heterogeneous nuclear ribonucleoprotein D0 | | Authors: | Enokizono, Y, Konishi, Y, Nagata, K, Ouhashi, K, Uesugi, S, Ishikawa, F, Katahira, M. | | Deposit date: | 2005-03-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D.
J.Biol.Chem., 280, 2005
|
|
