6NVQ
 
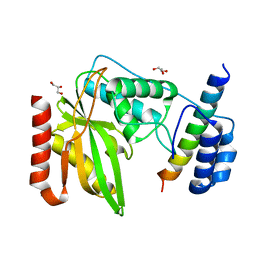 | |
6FPV
 
 | | A llama-derived JBP1-targeting nanobody | | 分子名称: | GLYCEROL, Nanobody | | 著者 | van Beusekom, B, Adamopoulos, A, Heidebrecht, T, Joosten, R.P, Perrakis, A. | | 登録日 | 2018-02-12 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Characterization and structure determination of a llama-derived nanobody targeting the J-base binding protein 1.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7Z5H
 
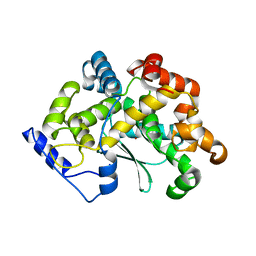 | | human Zn MATCAP | | 分子名称: | Uncharacterized protein KIAA0895-like, ZINC ION | | 著者 | Bak, J, Adamopoulos, A, Heidebrecht, T, Perrakis, A. | | 登録日 | 2022-03-09 | | 公開日 | 2022-05-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Posttranslational modification of microtubules by the MATCAP detyrosinase.
Science, 376, 2022
|
|
7Z5G
 
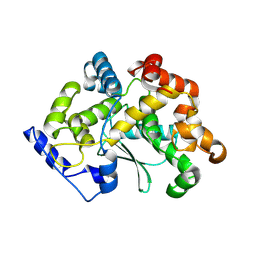 | |
7Z6S
 
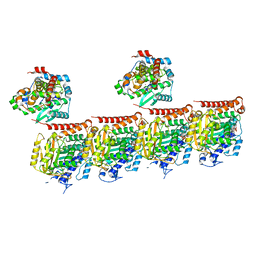 | |
7M6B
 
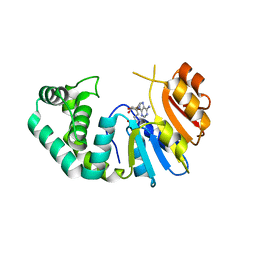 | | The Crystal Structure of Mcbe1 | | 分子名称: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, S-ADENOSYLMETHIONINE, ... | | 著者 | Alahuhta, P.M, Lunin, V.V. | | 登録日 | 2021-03-25 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Target highlights in CASP14: Analysis of models by structure providers.
Proteins, 89, 2021
|
|
7OC9
 
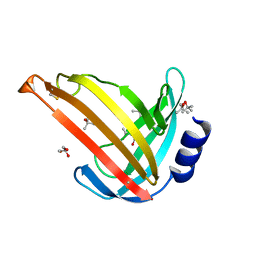 | |
7BG6
 
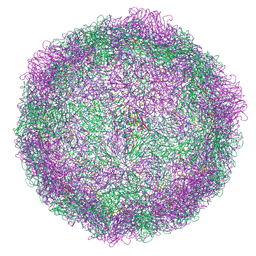 | | HRV14 native particle solved by cryoEM | | 分子名称: | Genome polyprotein, RNA-octamer (5'-R(P*UP*GP*UP*UP*UP*UP*UP*A)-3') | | 著者 | Hrebik, D, Fuzik, T, Plevka, P. | | 登録日 | 2021-01-06 | | 公開日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | ICAM-1 induced rearrangements of capsid and genome prime rhinovirus 14 for activation and uncoating.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BG7
 
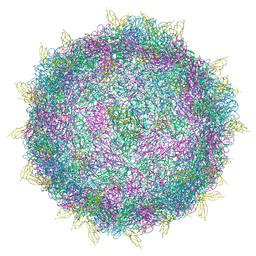 | |
6QLY
 
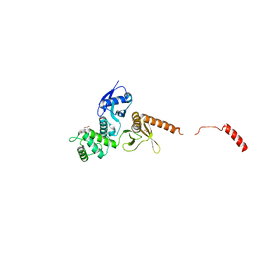 | | IDOL FERM domain | | 分子名称: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase MYLIP, SULFATE ION | | 著者 | Martinelli, L, Sixma, T.K. | | 登録日 | 2019-02-01 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural analysis of the LDL receptor-interacting FERM domain in the E3 ubiquitin ligase IDOL reveals an obscured substrate-binding site.
J.Biol.Chem., 295, 2020
|
|
6QLZ
 
 | | IDOL F3ab subdomain | | 分子名称: | E3 ubiquitin-protein ligase MYLIP | | 著者 | Martinelli, L, Johansson, P, Wan, P.T, Gunnarsson, J, Guo, H, Boyd, H. | | 登録日 | 2019-02-01 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.343 Å) | | 主引用文献 | Structural analysis of the LDL receptor-interacting FERM domain in the E3 ubiquitin ligase IDOL reveals an obscured substrate-binding site.
J.Biol.Chem., 295, 2020
|
|
7NUM
 
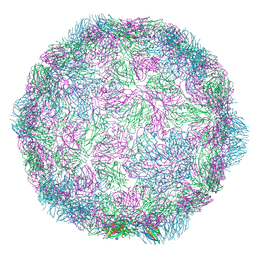 | |
7NUO
 
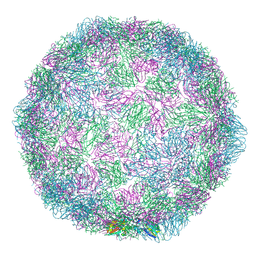 | |
7NUL
 
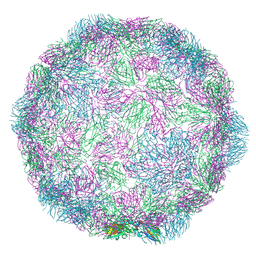 | |
7NUQ
 
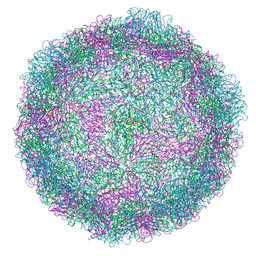 | | Rhinovirus 14 virion-like at pH 6.2 | | 分子名称: | Genome polyprotein, Octanucleotide | | 著者 | Hrebik, D, Plevka, P. | | 登録日 | 2021-03-12 | | 公開日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | ICAM-1 induced rearrangements of capsid and genome prime rhinovirus 14 for activation and uncoating.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NUN
 
 | |
