1V5C
 
 | | The crystal structure of the inactive form chitosanase from Bacillus sp. K17 at pH3.7 | | 分子名称: | SULFATE ION, chitosanase | | 著者 | Adachi, W, Shimizu, S, Sunami, T, Fukazawa, T, Suzuki, M, Yatsunami, R, Nakamura, S, Takenaka, A. | | 登録日 | 2003-11-22 | | 公開日 | 2004-12-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of family GH-8 chitosanase with subclass II specificity from Bacillus sp. K17
J.MOL.BIOL., 343, 2004
|
|
1V5D
 
 | | The crystal structure of the active form chitosanase from Bacillus sp. K17 at pH6.4 | | 分子名称: | PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), chitosanase | | 著者 | Adachi, W, Shimizu, S, Sunami, T, Fukazawa, T, Suzuki, M, Yatsunami, R, Nakamura, S, Takenaka, A. | | 登録日 | 2003-11-22 | | 公開日 | 2004-12-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of family GH-8 chitosanase with subclass II specificity from Bacillus sp. K17
J.MOL.BIOL., 343, 2004
|
|
1V59
 
 | | Crystal structure of yeast lipoamide dehydrogenase complexed with NAD+ | | 分子名称: | Dihydrolipoamide dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Adachi, W, Suzuki, K, Tsunoda, M, Sekiguchi, T, Reed, L.J, Takenaka, A. | | 登録日 | 2003-11-21 | | 公開日 | 2005-02-15 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of yeast lipoamide dehydrogenase complexed with NAD+
To be Published
|
|
2K6Q
 
 | | LC3 p62 complex structure | | 分子名称: | Microtubule-associated proteins 1A/1B light chain 3B, p62_peptide from Sequestosome-1 | | 著者 | Noda, N, Kumeta, H, Nakatogawa, H, Satoo, K, Adachi, W, Ishii, J, Fujioka, Y, Ohsumi, Y, Inagaki, F. | | 登録日 | 2008-07-17 | | 公開日 | 2008-09-02 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis of target recognition by ATG8/LC3 during selective autophagy
To be Published
|
|
2KWC
 
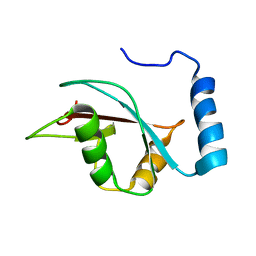 | | The NMR structure of the autophagy-related protein Atg8 | | 分子名称: | Autophagy-related protein 8 | | 著者 | Kumeta, H, Watanabe, M, Nakatogawa, H, Yamaguchi, M, Ogura, K, Adachi, W, Fujioka, Y, Noda, N.N, Ohsumi, Y, Inagaki, F. | | 登録日 | 2010-04-05 | | 公開日 | 2010-05-12 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The NMR structure of the autophagy-related protein Atg8
J.Biomol.Nmr, 47, 2010
|
|
5JH9
 
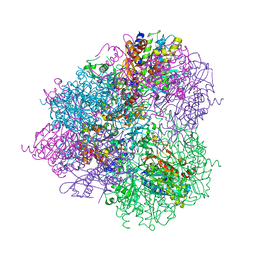 | | Crystal structure of prApe1 | | 分子名称: | CACODYLATE ION, Vacuolar aminopeptidase 1, ZINC ION | | 著者 | Noda, N.N, Adachi, W, Inagaki, F. | | 登録日 | 2016-04-20 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis for Receptor-Mediated Selective Autophagy of Aminopeptidase I Aggregates
Cell Rep, 16, 2016
|
|
5JGF
 
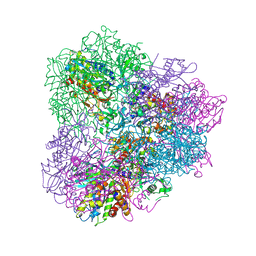 | | Crystal structure of mApe1 | | 分子名称: | Vacuolar aminopeptidase 1, ZINC ION | | 著者 | Noda, N.N, Adachi, W, Inagaki, F. | | 登録日 | 2016-04-20 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Structural Basis for Receptor-Mediated Selective Autophagy of Aminopeptidase I Aggregates
Cell Rep, 16, 2016
|
|
2YQU
 
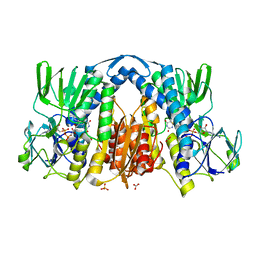 | | Crystal structures and evolutionary relationship of two different lipoamide dehydrogenase(E3s) from Thermus thermophilus | | 分子名称: | 2-oxoglutarate dehydrogenase E3 component, CARBONATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Kondo, H, Hossain, M.T, Adachi, W, Nakai, T, Kamiya, N, Kuramitsu, K. | | 登録日 | 2007-03-31 | | 公開日 | 2008-04-01 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structures and evolutionary relationship of two different lipoamide dehydrogenase(E3s) from Thermus thermophilus
To be Published
|
|
3VP7
 
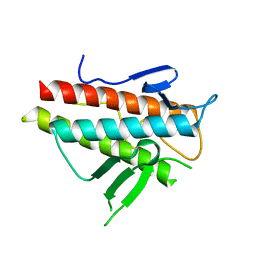 | | Crystal structure of the beta-alpha repeated, autophagy-specific (BARA) domain of Vps30/Atg6 | | 分子名称: | Vacuolar protein sorting-associated protein 30 | | 著者 | Noda, N.N, Kobayashi, T, Adachi, W, Fujioka, Y, Ohsumi, Y, Inagaki, F. | | 登録日 | 2012-02-28 | | 公開日 | 2012-03-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy.
J.Biol.Chem., 287, 2012
|
|
5JGE
 
 | |
5JHC
 
 | |
2ZPN
 
 | |
2N5L
 
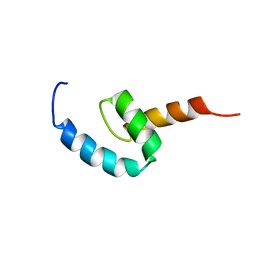 | | Regnase-1 C-terminal domain | | 分子名称: | Ribonuclease ZC3H12A | | 著者 | Yokogawa, M, Tsushima, T, Noda, N.N, Kumeta, H, Adachi, W, Enokizono, Y, Yamashita, K, Standley, D.M, Takeuchi, O, Akira, S, Inagaki, F. | | 登録日 | 2015-07-18 | | 公開日 | 2016-03-16 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
2N5J
 
 | | Regnase-1 N-terminal domain | | 分子名称: | Ribonuclease ZC3H12A | | 著者 | Yokogawa, M, Tsushima, T, Noda, N.N, Kumeta, H, Adachi, W, Enokizono, Y, Yamashita, K, Standley, D.M, Takeuchi, O, Akira, S, Inagaki, F. | | 登録日 | 2015-07-18 | | 公開日 | 2016-03-16 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
2N5K
 
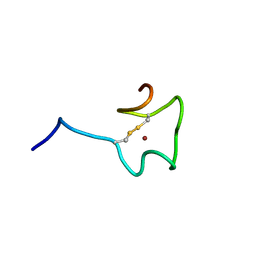 | | Regnase-1 Zinc finger domain | | 分子名称: | Ribonuclease ZC3H12A, ZINC ION | | 著者 | Yokogawa, M, Tsushima, T, Noda, N.N, Kumeta, H, Adachi, W, Enokizono, Y, Yamashita, K, Standley, D.M, Takeuchi, O, Akira, S, Inagaki, F. | | 登録日 | 2015-07-18 | | 公開日 | 2016-03-16 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
1V3O
 
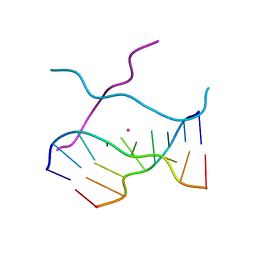 | | Crystal structure of d(GCGAGAGC): the DNA quadruplex structure split from the octaplex | | 分子名称: | 5'-D(*GP*(C38)P*GP*AP*GP*AP*GP*C)-3', POTASSIUM ION | | 著者 | Kondo, J, Umeda, S, Sunami, T, Takenaka, A. | | 登録日 | 2003-11-03 | | 公開日 | 2004-06-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structures of a DNA octaplex with I-motif of G-quartets and its splitting into two quadruplexes suggest a folding mechanism of eight tandem repeats
Nucleic Acids Res., 32, 2004
|
|
1V3N
 
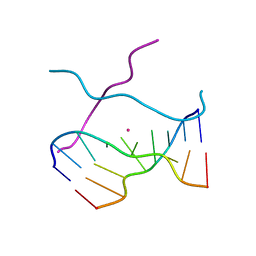 | | Crystal structure of d(GCGAGAGC): the DNA quadruplex structure split from the octaplex | | 分子名称: | 5'-D(*GP*(CBR)P*GP*AP*GP*AP*GP*C)-3', POTASSIUM ION | | 著者 | Kondo, J, Umeda, S, Sunami, T, Takenaka, A. | | 登録日 | 2003-11-03 | | 公開日 | 2004-06-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of a DNA octaplex with I-motif of G-quartets and its splitting into two quadruplexes suggest a folding mechanism of eight tandem repeats
Nucleic Acids Res., 32, 2004
|
|
1V3P
 
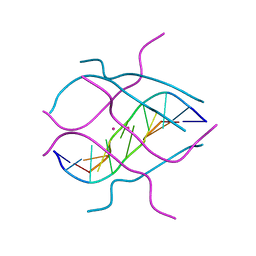 | | Crystal structure of d(GCGAGAGC): the DNA octaplex structure with I-motif of G-quartet | | 分子名称: | 5'-D(*GP*(C38)P*GP*AP*GP*AP*GP*C)-3', POTASSIUM ION | | 著者 | Kondo, J, Umeda, S, Sunami, T, Takenaka, A. | | 登録日 | 2003-11-03 | | 公開日 | 2004-06-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of a DNA octaplex with I-motif of G-quartets and its splitting into two quadruplexes suggest a folding mechanism of eight tandem repeats
Nucleic Acids Res., 32, 2004
|
|
1V5B
 
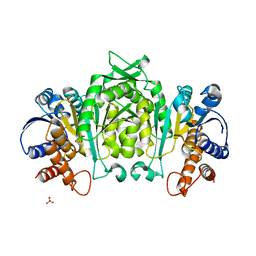 | | The Structure Of The Mutant, S225A and E251L, Of 3-Isopropylmalate Dehydrogenase From Bacillus Coagulans | | 分子名称: | 3-isopropylmalate dehydrogenase, SULFATE ION | | 著者 | Fujita, K, Minami, H, Suzuki, K, Tsunoda, M, Sekiguchi, T, Mizui, R, Tsuzaki, S, Nakamura, S, Takenaka, A. | | 登録日 | 2003-11-22 | | 公開日 | 2005-02-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Crystal structure of a highly thermo-stabilized mutant of 3-isopropylmalate dehydrogenase from Bacillus coagulans: An evaluation of local packing density in the hydrophobic core
To be Published
|
|
5H9V
 
 | | Crystal structure of Regnase PIN domain, form I | | 分子名称: | Ribonuclease ZC3H12A, SODIUM ION | | 著者 | Yokogawa, M, Tsushima, T, Adachi, W, Noda, N.N, Inagaki, F. | | 登録日 | 2015-12-29 | | 公開日 | 2016-03-16 | | 最終更新日 | 2020-02-19 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
5H9W
 
 | | Crystal structure of Regnase PIN domain, form II | | 分子名称: | Ribonuclease ZC3H12A, SODIUM ION | | 著者 | Yokogawa, M, Tsushima, T, Adachi, W, Noda, N.N, Inagaki, F. | | 登録日 | 2015-12-29 | | 公開日 | 2016-03-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
