3KDE
 
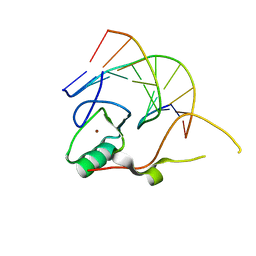 | | Crystal structure of the THAP domain from D. melanogaster P-element transposase in complex with its natural DNA binding site | | Descriptor: | 5'-D(*(BRU)P*CP*CP*AP*CP*TP*TP*AP*AP*C)-3', 5'-D(*GP*TP*TP*AP*AP*GP*(BRU)P*GP*GP*A)-3', Transposable element P transposase, ... | | Authors: | Sabogal, A, Lyubimov, A.Y, Berger, J.M, Rio, D.C. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | THAP proteins target specific DNA sites through bipartite recognition of adjacent major and minor grooves.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7O3M
 
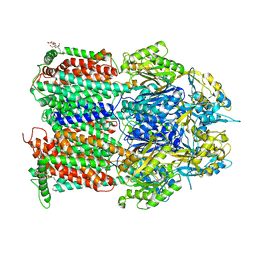 | | Crystal Structure of AcrB Single Mutant - 1 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Efflux pump membrane transporter | | Authors: | Ababou, A. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.551 Å) | | Cite: | Crystal Structure of AcrB Single Mutant - 1
To Be Published
|
|
7O3L
 
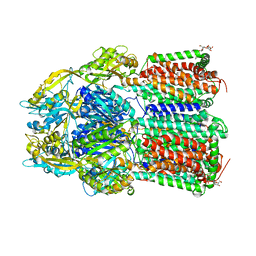 | | Crystal Structure of AcrB Double Mutant | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Efflux pump membrane transporter | | Authors: | Ababou, A. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.526 Å) | | Cite: | Crystal Structure of AcrB Double Mutant
To Be Published
|
|
7O3N
 
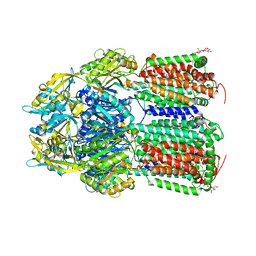 | | Crystal Structure of AcrB Single Mutant - 2 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Efflux pump membrane transporter | | Authors: | Ababou, A. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.561 Å) | | Cite: | Crystal Structure of AcrB Single Mutant - 2
To Be Published
|
|
5JGH
 
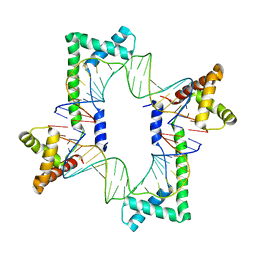 | | Crystal structure of the mitochondrial DNA packaging protein Abf2p in complex with DNA at 2.6 Angstrom resolution | | Descriptor: | ACETATE ION, ARS-binding factor 2, mitochondrial, ... | | Authors: | Chakraborty, A, Lyonnais, S, Sola, M. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DNA structure directs positioning of the mitochondrial genome packaging protein Abf2p.
Nucleic Acids Res., 45, 2017
|
|
5DSU
 
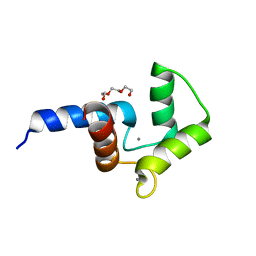 | | Crystal structure of double mutant of N-domain of human calmodulin | | Descriptor: | CALCIUM ION, Calmodulin, TRIETHYLENE GLYCOL | | Authors: | Ababou, A, Zaleska, M. | | Deposit date: | 2015-09-17 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | On the Ca(2+) binding and conformational change in EF-hand domains: Experimental evidence of Ca(2+)-saturated intermediates of N-domain of calmodulin.
Biochim. Biophys. Acta, 1865, 2017
|
|
4EZ2
 
 | |
2I08
 
 | |
4BPZ
 
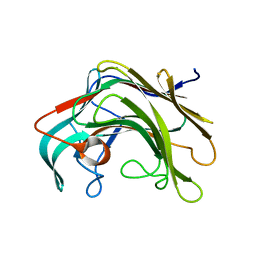 | | Crystal structure of lamA_E269S from Zobellia galactanivorans in complex with a trisaccharide of 1,3-1,4-beta-D-glucan. | | Descriptor: | CALCIUM ION, ENDO-1,3-BETA-GLUCANASE, FAMILY GH16, ... | | Authors: | Labourel, A, Jam, M, Jeudy, A, Czjzek, M, Michel, G. | | Deposit date: | 2013-05-29 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | The Beta-Glucanase Zglama from Zobellia Galactanivorans Evolved a Bent Active Site Adapted for Efficient Degradation of Algal Laminarin.
J.Biol.Chem., 289, 2014
|
|
4BQ1
 
 | | Crystal structure of of LamAcat from Zobellia galactanivorans | | Descriptor: | CALCIUM ION, ENDO-1,3-BETA-GLUCANASE, FAMILY GH16, ... | | Authors: | Labourel, A, Jam, M, Jeudy, A, Michel, G, Czjzek, M. | | Deposit date: | 2013-05-29 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Beta Glucanase Zglama from Zobellia Galactanivorans Evolved a Bent Active Site Adapted for Efficient Degradation of Algal Laminarin
J.Biol.Chem., 289, 2014
|
|
4BOW
 
 | | Crystal structure of LamA_E269S from Z. galactanivorans in complex with laminaritriose and laminaritetraose | | Descriptor: | CALCIUM ION, ENDO-1,3-BETA-GLUCANASE, FAMILY GH16, ... | | Authors: | Labourel, A, Jeudy, A, Czjzek, M, Michel, G. | | Deposit date: | 2013-05-22 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Beta-Glucanase Zglama from Zobellia Galactanivorans Evolved a Bent Active Site Adapted for Efficient Degradation of Algal Laminarin
J.Biol.Chem., 289, 2014
|
|
2AVG
 
 | |
4CRQ
 
 | | Crystal structure of the catalytic domain of the modular laminarinase ZgLamC mutant E142S | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Labourel, A, Jam, M, Legentil, L, Sylla, B, Ficko-Blean, E, Hehemann, J.H, Ferrieres, V, Czjzek, M, Michel, G. | | Deposit date: | 2014-02-28 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Biochemical Characterization of the Laminarina Zglamc[Gh16] from Zobellia Galactanivorans Suggests Preferred Recognition of Branched Laminarin
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1PD6
 
 | |
4CTE
 
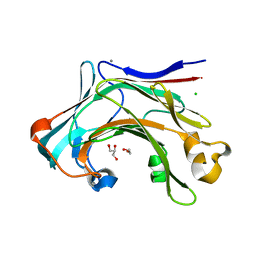 | | Crystal structure of the catalytic domain of the modular laminarinase ZgLamC mutant E142S in complex with a thio-oligosaccharide | | Descriptor: | 1,2-ETHANEDIOL, 1-thio-beta-D-glucopyranose-(1-3)-1-thio-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Labourel, A, Jam, M, Legentil, L, Sylla, B, Ficko-Blean, E, Hehemann, J.H, Ferrieres, V, Czjzek, M, Michel, G. | | Deposit date: | 2014-03-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Characterization of the Laminarina Zglamc[Gh16] from Zobellia Galactanivorans Suggests Preferred Recognition of Branched Laminarin
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2O2O
 
 | |
8G8D
 
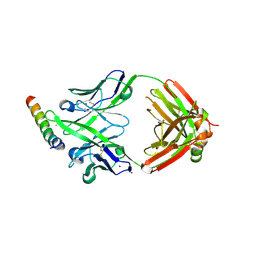 | | Crystal structure of DH1346 Fab in complex with HIV proximal MPER peptide | | Descriptor: | DH1346 heavy chain, DH1346 light chain, FLUORIDE ION, ... | | Authors: | Niyongabo, A, Janus, B.M, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
8G8C
 
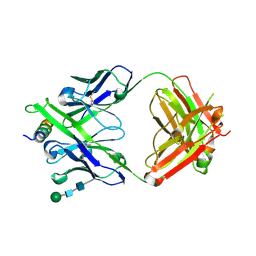 | | Crystal structure of DH1322.1 Fab in complex with HIV proximal MPER peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1322.1 heavy chain, DH1322.1 light chain, ... | | Authors: | Niyongabo, A, Janus, B.M, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
5LA1
 
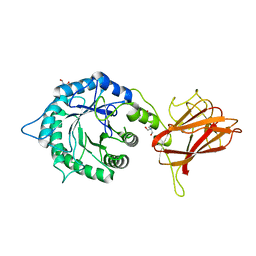 | | The mechanism by which arabinoxylanases can recognise highly decorated xylans | | Descriptor: | CALCIUM ION, Carbohydrate binding family 6, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, ... | | Authors: | Basle, A, Labourel, A, Cuskin, F, Jackson, A, Crouch, L, Rogowski, A, Gilbert, H. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Mechanism by Which Arabinoxylanases Can Recognize Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
5LA2
 
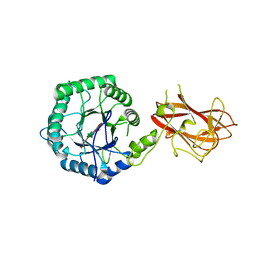 | | The mechanism by which arabinoxylanases can recognise highly decorated xylans | | Descriptor: | CALCIUM ION, Carbohydrate binding family 6, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-[alpha-L-arabinofuranose-(1-3)]alpha-D-xylopyranose, ... | | Authors: | Basle, A, Labourel, A, Cuskin, F, Jackson, A, Crouch, L, Rogowski, A, Gilbert, H. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Mechanism by Which Arabinoxylanases Can Recognize Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
5LA0
 
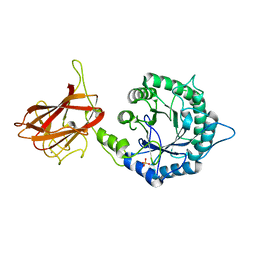 | | The mechanism by which arabinoxylanases can recognise highly decorated xylans | | Descriptor: | CALCIUM ION, Carbohydrate binding family 6, SULFATE ION, ... | | Authors: | Basle, A, Labourel, A, Cuskin, F, Jackson, A, Crouch, L, Rogowski, A, Gilbert, A. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Mechanism by Which Arabinoxylanases Can Recognize Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
6HZF
 
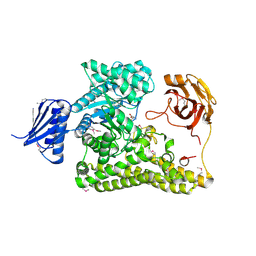 | | BP0997, GH138 enzyme targeting pectin rhamnogalacturonan II | | Descriptor: | BPa0997, SERINE, SODIUM ION | | Authors: | Basle, A, Cartmell, A, Labourel, A, Gilbert, H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and functional analyses of glycoside hydrolase 138 enzymes targeting chain A galacturonic acid in the complex pectin rhamnogalacturonan II.
J.Biol.Chem., 294, 2019
|
|
6IBH
 
 | | Copper binding protein from Laetisaria arvalis (LaX325) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity CAZyme, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Tandrup, T, Labourel, A, Haon, M, Berrin, J.-G, Lo Leggio, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-11-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A fungal family of lytic polysaccharide monooxygenase-like copper proteins.
Nat.Chem.Biol., 16, 2020
|
|
6IBI
 
 | | Copper binding protein from Laetisaria arvalis (LaX325) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity CAZyme, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Tandrup, T, Labourel, A, Haon, M, Berrin, J.-G, Lo Leggio, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A fungal family of lytic polysaccharide monooxygenase-like copper proteins.
Nat.Chem.Biol., 16, 2020
|
|
6HZE
 
 | | BP0997, GH138 enzyme targeting pectin rhamnogalacturonan II | | Descriptor: | BPa0997, SODIUM ION, beta-D-galactopyranuronic acid | | Authors: | Basle, A, Cartmell, A, Labourel, A, Gilbert, H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional analyses of glycoside hydrolase 138 enzymes targeting chain A galacturonic acid in the complex pectin rhamnogalacturonan II.
J.Biol.Chem., 294, 2019
|
|
