4XBY
 
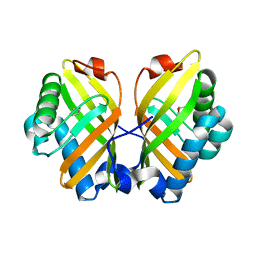 | | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH complexed with cyclopentene oxide | | Descriptor: | (1R,5S)-6-oxabicyclo[3.1.0]hexane, Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XDW
 
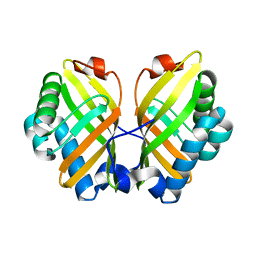 | | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH | | Descriptor: | Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Lonsdale, R, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-20 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1QSR
 
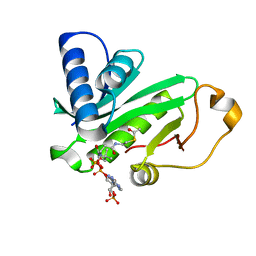 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 WITH BOUND ACETYL-COENZYME A | | Descriptor: | ACETYL COENZYME *A, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
1QST
 
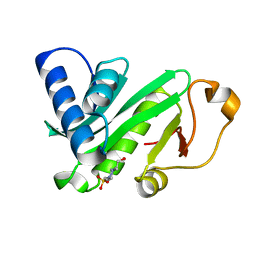 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
1QSN
 
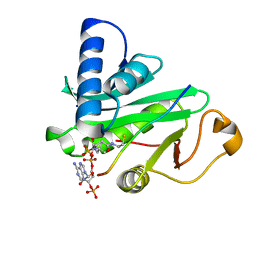 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 WITH BOUND COENZYME A AND HISTONE H3 PEPTIDE | | Descriptor: | COENZYME A, HISTONE H3, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-22 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
4JCL
 
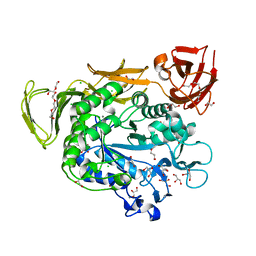 | | Crystal structure of Alpha-CGT from Paenibacillus macerans at 1.7 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J, Wu, J, Li, J, Chen, J. | | Deposit date: | 2013-02-22 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Alpha-Cgt from Paenibacillus Macerans at 1.7 Angstrom Resolution
To be Published
|
|
4P5A
 
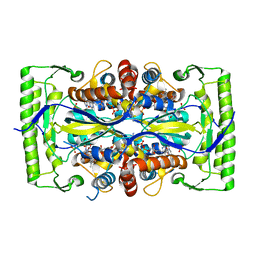 | | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi bound with 5-Br UMP | | Descriptor: | 5-BROMO-URIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase ThyX | | Authors: | Li, Y, Chen, W, Li, J, Xia, Z, Deng, Z, Zhou, J. | | Deposit date: | 2014-03-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi with 5-Br UMP
To Be Published
|
|
4P5B
 
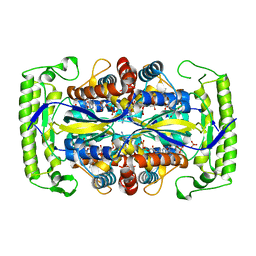 | | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi bound with 5-Br dUMP | | Descriptor: | 5-BROMO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Li, Y, Chen, W, Li, J, Xia, Z, Deng, Z, Zhou, J. | | Deposit date: | 2014-03-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.274 Å) | | Cite: | Crystal structure of a UMP/dUMP methylase PolB form Streptomyces cacaoi bound with 5-Br dUMP
To Be Published
|
|
2K96
 
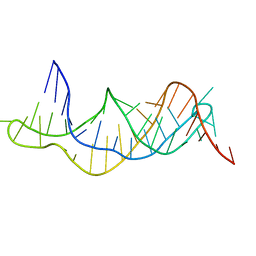 | | Solution structure of the RDC-refined P2B-P3 pseudoknot from human telomerase RNA (delta U177) | | Descriptor: | TELOMERASE RNA P2B-P3 PSEUDOKNOT | | Authors: | Kim, N.-K, Zhang, Q, Zhou, J, Theimer, C.A, Peterson, R.D, Feigon, J. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of the Wild-type Pseudoknot of Human Telomerase RNA.
J.Mol.Biol., 384, 2008
|
|
4LJY
 
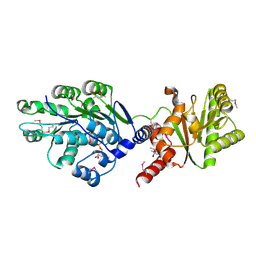 | | Crystal structure of RNA splicing effector Prp5 in complex with ADP | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhang, Z.-M, Li, J, Yang, F, Xu, Y, Zhou, J. | | Deposit date: | 2013-07-05 | | Release date: | 2013-12-11 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Prp5p reveals interdomain interactions that impact spliceosome assembly.
Cell Rep, 5, 2013
|
|
4LK2
 
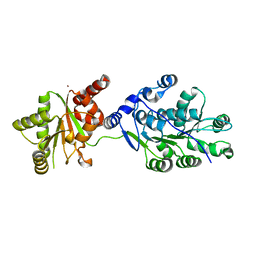 | | Crystal structure of RNA splicing effector Prp5 | | Descriptor: | NICKEL (II) ION, Pre-mRNA-processing ATP-dependent RNA helicase PRP5 | | Authors: | Zhang, Z.-M, Li, J, Yang, F, Xu, Y, Zhou, J. | | Deposit date: | 2013-07-05 | | Release date: | 2013-12-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of Prp5p reveals interdomain interactions that impact spliceosome assembly.
Cell Rep, 5, 2013
|
|
6JJ8
 
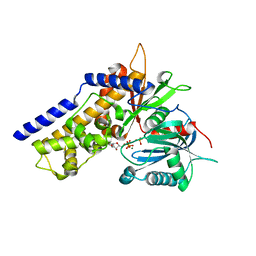 | | Crystal structure of OsHXK6-ATP-Mg2+ complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of OsHXK6-ATP-Mg2+ complex
To Be Published
|
|
6JJ7
 
 | | Crystal structure of OsHXK6-Glc complex | | Descriptor: | Rice hexokinase 6, beta-D-glucopyranose | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of OsHXK6-Glc complex
To Be Published
|
|
6JJ4
 
 | | Crystal structure of OsHXK6-apo form | | Descriptor: | Hexokinase-6 | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of OsHXK6-apo
To Be Published
|
|
6JJ9
 
 | | Crystal structure of OsHXK6-Glc-ATP-Mg2+ complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Hexokinase-6, MAGNESIUM ION, ... | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of OsHXK6-Glc-ATP-Mg2+ complex
To Be Published
|
|
3H5W
 
 | | Crystal structure of the GluR2-ATD in space group P212121 without solvent | | Descriptor: | Glutamate receptor 2 | | Authors: | Jin, R, Singh, S.K, Gu, S, Furukawa, H, Sobolevsky, A, Zhou, J, Jin, Y, Gouaux, E. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | Crystal structure and association behaviour of the GluR2 amino-terminal domain.
Embo J., 28, 2009
|
|
3H5V
 
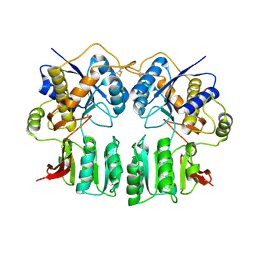 | | Crystal structure of the GluR2-ATD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2 | | Authors: | Jin, R, Singh, S.K, Gu, S, Furukawa, H, Sobolevsky, A, Zhou, J, Jin, Y, Gouaux, E. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure and association behaviour of the GluR2 amino-terminal domain.
Embo J., 28, 2009
|
|
8JLV
 
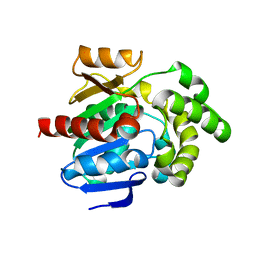 | |
8JNX
 
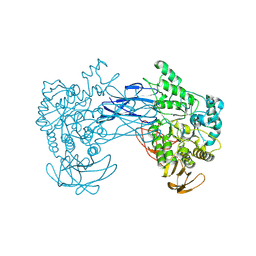 | |
8JN0
 
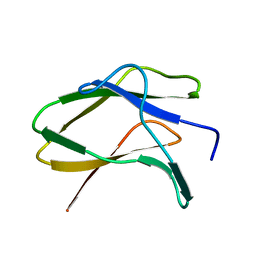 | | N/F domain of alkaline amylase Amy703 | | Descriptor: | Alpha-amylase | | Authors: | Xiang, L, Zhang, G, Zhou, J. | | Deposit date: | 2023-06-05 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.22826374 Å) | | Cite: | Truncation of N-terminus domain of alkaline a-amylase to form a unique dimer leads to improved activity and stability and decreased calcium ion dependence
To Be Published
|
|
4JCM
 
 | | Crystal structure of Gamma-CGTASE from Alkalophilic bacillus clarkii at 1.65 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, L, Yang, D, Zhou, J, Wu, J, Chen, J. | | Deposit date: | 2013-02-22 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of Gamma-Cgtase from Alkalophilic Bacillus Clarkii at 1.65 Angstrom Resolution.
To be Published
|
|
5C54
 
 | |
5C55
 
 | |
7RST
 
 | | The Crystal Structure of Recombinant Chloroperoxidase Expressed in Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chloroperoxidase, ... | | Authors: | Tang, X, Venkadesh, S, Zhou, J, Rosen, B, Wang, X. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The Crystal Structure of Recombinant Chloroperoxidase Expressed in Aspergillus niger
To Be Published
|
|
2K95
 
 | | Solution structure of the wild-type P2B-P3 pseudoknot of human telomerase RNA | | Descriptor: | Telomerase RNA P2b-P3 pseudoknot | | Authors: | Kim, N.-K, Zhang, Q, Zhou, J, Theimer, C.A, Peterson, R.D, Feigon, J. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of the Wild-type Pseudoknot of Human Telomerase RNA.
J.Mol.Biol., 384, 2008
|
|
