3FIS
 
 | | THE MOLECULAR STRUCTURE OF WILD-TYPE AND A MUTANT FIS PROTEIN: RELATIONSHIP BETWEEN MUTATIONAL CHANGES AND RECOMBINATIONAL ENHANCER FUNCTION OR DNA BINDING | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS) | | Authors: | Yuan, H.S, Finkel, S.E, Feng, J-A, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 1991-08-12 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular structure of wild-type and a mutant Fis protein: relationship between mutational changes and recombinational enhancer function or DNA binding.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
4FIS
 
 | | THE MOLECULAR STRUCTURE OF WILD-TYPE AND A MUTANT FIS PROTEIN: RELATIONSHIP BETWEEN MUTATIONAL CHANGES AND RECOMBINATIONAL ENHANCER FUNCTION OR DNA BINDING | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS) | | Authors: | Yuan, H.S, Finkel, S.E, Feng, J.-A, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 1991-08-12 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular structure of wild-type and a mutant Fis protein: relationship between mutational changes and recombinational enhancer function or DNA binding.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1FIP
 
 | | THE STRUCTURE OF FIS MUTANT PRO61ALA ILLUSTRATES THAT THE KINK WITHIN THE LONG ALPHA-HELIX IS NOT DUE TO THE PRESENCE OF THE PROLINE RESIDUE | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS), UNKNOWN PEPTIDE, POSSIBLY PART OF THE UNOBSERVED RESIDUES IN ENTITY 1 | | Authors: | Yuan, H.S, Wang, S.S, Yang, W.-Z, Finkel, S.E, Johnson, R.C. | | Deposit date: | 1994-09-26 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of Fis mutant Pro61Ala illustrates that the kink within the long alpha-helix is not due to the presence of the proline residue.
J.Biol.Chem., 269, 1994
|
|
1OUO
 
 | |
1OUP
 
 | |
3S5B
 
 | |
5ZF6
 
 | | Crystal structure of the dimeric human PNPase | | Descriptor: | Polyribonucleotide nucleotidyltransferase 1, mitochondrial | | Authors: | Yuan, H.S, Golzarroshan, B. | | Deposit date: | 2018-03-02 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Crystal structure of dimeric human PNPase reveals why disease-linked mutants suffer from low RNA import and degradation activities.
Nucleic Acids Res., 46, 2018
|
|
1M08
 
 | | Crystal structure of the unbound nuclease domain of ColE7 | | Descriptor: | Colicin E7, PHOSPHATE ION, ZINC ION | | Authors: | Cheng, Y.S, Hsia, K.C, Doudeva, L.G, Chak, K.F, Yuan, H.S. | | Deposit date: | 2002-06-12 | | Release date: | 2002-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Nuclease Domain of Colicin E7 Suggests a Mechanism for Binding to Double-stranded DNA by the H-N-H Endonucleases
J.mol.biol., 324, 2002
|
|
3KRN
 
 | | Crystal Structure of C. elegans cell-death-related nuclease 5(CRN-5) | | Descriptor: | Protein C14A4.5, confirmed by transcript evidence | | Authors: | Yang, C.-C, Wang, Y.-T, Hsiao, Y.-Y, Doudeva, L.G, Chow, S.Y, Yuan, H.S. | | Deposit date: | 2009-11-19 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.918 Å) | | Cite: | Structural and biochemical characterization of CRN-5 and Rrp46: an exosome component participating in apoptotic DNA degradation
Rna, 16, 2010
|
|
3CDJ
 
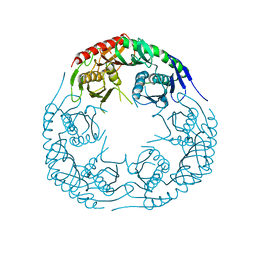 | | Crystal structure of the E. coli KH/S1 domain truncated PNPase | | Descriptor: | Polynucleotide phosphorylase | | Authors: | Shi, Z, Yang, W.Z, Lin-Chao, S, Chak, K.F, Yuan, H.S. | | Deposit date: | 2008-02-27 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive RNA degradation.
Rna, 14, 2008
|
|
3CDI
 
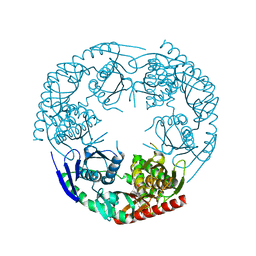 | | Crystal structure of E. coli PNPase | | Descriptor: | Polynucleotide phosphorylase | | Authors: | Shi, Z, Yang, W.Z, Lin-Chao, S, Chak, K.F, Yuan, H.S. | | Deposit date: | 2008-02-27 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive RNA degradation.
Rna, 14, 2008
|
|
3CG7
 
 | |
3TAT
 
 | | TYROSINE AMINOTRANSFERASE FROM E. COLI | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, TYROSINE AMINOTRANSFERASE | | Authors: | Ko, T.P, Yang, W.Z, Wu, S.P, Tsai, H, Yuan, H.S. | | Deposit date: | 1998-08-12 | | Release date: | 1999-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallization and preliminary crystallographic analysis of the Escherichia coli tyrosine aminotransferase.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2AXC
 
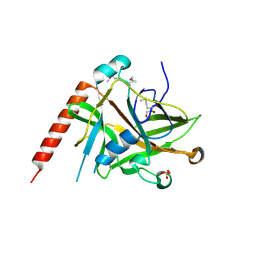 | | Crystal structure of ColE7 translocation domain | | Descriptor: | Colicin E7, GLYCEROL, SULFATE ION | | Authors: | Cheng, Y.S, Shi, Z, Doudeva, L.G, Yang, W.Z, Chak, K.F, Yuan, H.S. | | Deposit date: | 2005-09-04 | | Release date: | 2006-03-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structure of a truncated ColE7 translocation domain: implications for colicin transport across membranes
J.Mol.Biol., 356, 2006
|
|
4QN0
 
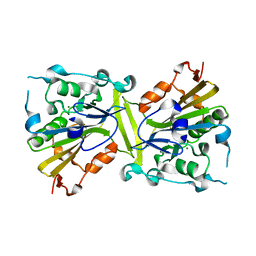 | | Crystal structure of the CPS-6 mutant Q130K | | Descriptor: | Endonuclease G, mitochondrial, MAGNESIUM ION | | Authors: | Lin, J.L.J, Yuan, H.S. | | Deposit date: | 2014-06-17 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Oxidative Stress Impairs Cell Death by Repressing the Nuclease Activity of Mitochondrial Endonuclease G
Cell Rep, 16, 2016
|
|
1UNK
 
 | | STRUCTURE OF COLICIN E7 IMMUNITY PROTEIN | | Descriptor: | COLICIN E7 | | Authors: | Ko, T.-P, Hsieh, S.-Y, Ku, W.-Y, Tseng, M.-Y, Chak, K.-F, Yuan, H.S. | | Deposit date: | 1996-06-21 | | Release date: | 1998-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel role of ImmE7 in the autoregulatory expression of the ColE7 operon and identification of possible RNase active sites in the crystal structure of dimeric ImmE7.
EMBO J., 16, 1997
|
|
1MVE
 
 | | Crystal structure of a natural circularly-permutated jellyroll protein: 1,3-1,4-beta-D-glucanase from Fibrobacter succinogenes | | Descriptor: | CALCIUM ION, Truncated 1,3-1,4-beta-D-glucanase | | Authors: | Tsai, L.-C, Shyur, L.-F, Lee, S.-H, Lin, S.-S, Yuan, H.S. | | Deposit date: | 2002-09-25 | | Release date: | 2003-07-15 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Natural Circularly Permuted Jellyroll Protein: 1,3-1,4-beta-D-Glucanase from Fibrobacter succinogenes.
J.Mol.Biol., 330, 2003
|
|
1MZ8
 
 | | CRYSTAL STRUCTURES OF THE NUCLEASE DOMAIN OF COLE7/IM7 IN COMPLEX WITH A PHOSPHATE ION AND A ZINC ION | | Descriptor: | Colicin E7, Colicin E7 immunity protein, PHOSPHATE ION, ... | | Authors: | Sui, M.J, Tsai, L.C, Hsia, K.C, Doudeva, L.G, Ku, W.Y, Han, G.W, Yuan, H.S. | | Deposit date: | 2002-10-07 | | Release date: | 2002-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal ions and phosphate binding in the H-N-H motif: crystal structures of the nuclease domain of ColE7/Im7 in complex with a phosphate ion and different divalent metal ions
PROTEIN SCI., 11, 2002
|
|
1PT3
 
 | | Crystal structures of nuclease-ColE7 complexed with octamer DNA | | Descriptor: | 5'-GCGATCGC-3', Colicin E7 | | Authors: | Hsia, K.C, Chak, K.F, Cheng, Y.S, Ku, W.Y, Yuan, H.S. | | Deposit date: | 2003-06-22 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA binding and degradation by the HNH protein ColE7.
STRUCTURE, 12, 2004
|
|
3FBD
 
 | |
3CM6
 
 | |
3D2W
 
 | | Crystal structure of mouse TDP-43 RRM2 domain in complex with DNA | | Descriptor: | DNA (5'-D(*DGP*DTP*DTP*DGP*DAP*DGP*DCP*DGP*DTP*DT)-3'), PHOSPHATE ION, TAR DNA-binding protein 43 | | Authors: | Kuo, P.H, Yuan, H.S. | | Deposit date: | 2008-05-09 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into TDP-43 in nucleic-acid binding and domain interactions
Nucleic Acids Res., 37, 2009
|
|
3CM5
 
 | |
4Y00
 
 | | Crystal Structure of Human TDP-43 RRM1 Domain with D169G Mutation in Complex with an Unmodified Single-stranded DNA | | Descriptor: | DNA (5'-D(P*TP*TP*GP*AP*GP*CP*GP*T)-3'), TAR DNA-binding protein 43 | | Authors: | Chiang, C.H, Kuo, P.H, Yang, W.Z, Yuan, H.S. | | Deposit date: | 2015-02-05 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of disease-related TDP-43 D169G mutation: linking enhanced stability and caspase cleavage efficiency to protein accumulation
Sci Rep, 6, 2016
|
|
4Y0F
 
 | | Crystal Structure of Human TDP-43 RRM1 Domain in Complex with an Unmodified Single-stranded DNA | | Descriptor: | DNA (5'-D(*GP*TP*TP*GP*AP*GP*CP*GP*TP*T)-3'), TAR DNA-binding protein 43 | | Authors: | Chiang, C.H, Kuo, P.H, Doudeva, L.G, Wang, Y.T, Yuan, H.S. | | Deposit date: | 2015-02-06 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Structural analysis of disease-related TDP-43 D169G mutation: linking enhanced stability and caspase cleavage efficiency to protein accumulation
Sci Rep, 6, 2016
|
|
