7CCY
 
 | | Crystal structure of the 2-iodoporphobilinogen-bound holo form of human hydroxymethylbilane synthase | | Descriptor: | 3-[5-(aminomethyl)-4-(carboxymethyl)-2-iodo-1H-pyrrol-3-yl]propanoic acid, 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Sato, H, Sugishima, M, Wada, K, Hirabayashi, K, Tsukaguchi, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of hydroxymethylbilane synthase complexed with a substrate analog: a single substrate-binding site for four consecutive condensation steps.
Biochem.J., 478, 2021
|
|
2D1V
 
 | | Crystal structure of DNA-binding domain of Bacillus subtilis YycF | | Descriptor: | Transcriptional regulatory protein yycF | | Authors: | Okajima, T, Okada, A, Watanabe, T, Yamamoto, K, Tanizawa, K, Utsumi, R. | | Deposit date: | 2005-09-01 | | Release date: | 2006-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Response regulator YycF essential for bacterial growth: X-ray crystal structure of the DNA-binding domain and its PhoB-like DNA recognition motif
Febs Lett., 582, 2008
|
|
3WEX
 
 | | Crystal structure of HLA-DP5 in complex with Cry j 1-derived peptide (residues 214-222) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class II antigen | | Authors: | Kusano, S, Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-16 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the specific recognition of the major antigenic peptide from the Japanese cedar pollen allergen Cry j 1 by HLA-DP5
J. Mol. Biol., 426, 2014
|
|
6KME
 
 | | Crystal structure of phytochromobilin synthase from tomato in complex with biliverdin | | Descriptor: | BILIVERDINE IX ALPHA, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sugishima, M, Wada, K, Fukuyama, K. | | Deposit date: | 2019-07-31 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of phytochromobilin synthase in complex with biliverdin IX alpha , a key enzyme in the biosynthesis of phytochrome.
J.Biol.Chem., 295, 2020
|
|
6KMD
 
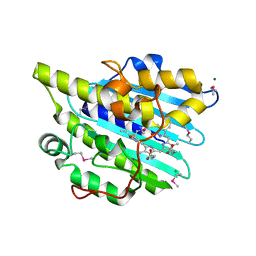 | | Crystal structure of SeMet-phytochromobilin synthase from tomato in complex with biliverdin | | Descriptor: | BILIVERDINE IX ALPHA, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sugishima, M, Wada, K, Fukuyama, K. | | Deposit date: | 2019-07-31 | | Release date: | 2019-12-18 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of phytochromobilin synthase in complex with biliverdin IX alpha , a key enzyme in the biosynthesis of phytochrome.
J.Biol.Chem., 295, 2020
|
|
8IBR
 
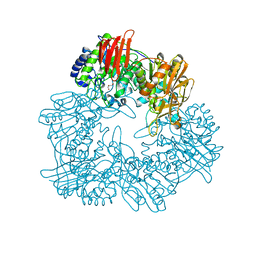 | | Crystal structure of GH42 beta-galactosidase BiBga42A from Bifidobacterium longum subspecies infantis in complex with glycerol | | Descriptor: | Beta-galactosidase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Hidaka, M, Fushinobu, S, Gotoh, A, Katayama, T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate recognition mode of a glycoside hydrolase family 42 beta-galactosidase from Bifidobacterium longum subspecies infantis ( Bi Bga42A) revealed by crystallographic and mutational analyses.
Microbiome Res Rep, 2, 2023
|
|
8IBT
 
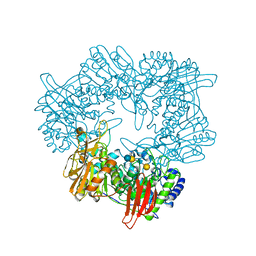 | | Crystal structure of GH42 beta-galactosidase BiBga42A from Bifidobacterium longum subspecies infantis E318S mutant in complex with lacto-N-tetraose | | Descriptor: | Beta-galactosidase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hidaka, M, Fushinobu, S, Gotoh, A, Katayama, T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate recognition mode of a glycoside hydrolase family 42 beta-galactosidase from Bifidobacterium longum subspecies infantis ( Bi Bga42A) revealed by crystallographic and mutational analyses.
Microbiome Res Rep, 2, 2023
|
|
8IBS
 
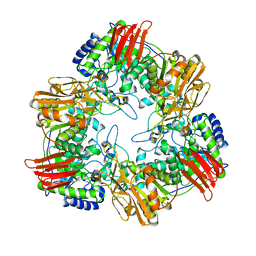 | | Crystal structure of GH42 beta-galactosidase BiBga42A from Bifidobacterium longum subspecies infantis E160A/E318A mutant in complex with galactose | | Descriptor: | Beta-galactosidase, alpha-D-galactopyranose | | Authors: | Hidaka, M, Fushinobu, S, Gotoh, A, Katayama, T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate recognition mode of a glycoside hydrolase family 42 beta-galactosidase from Bifidobacterium longum subspecies infantis ( Bi Bga42A) revealed by crystallographic and mutational analyses.
Microbiome Res Rep, 2, 2023
|
|
6J79
 
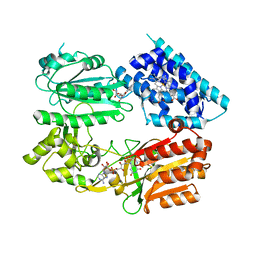 | | Fusion protein of heme oxygenase-1 and NADPH-cytochrome P450 reductase (13aa) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Heme oxygenase 1,NADPH--cytochrome P450 reductase, ... | | Authors: | Sugishima, M, Wada, K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.331 Å) | | Cite: | Crystal structure of a NADPH-cytochrome P450 oxidoreductase (CYPOR) and heme oxygenase 1 fusion protein implies a conformational change in CYPOR upon NADPH/NADP+binding.
Febs Lett., 593, 2019
|
|
1DBN
 
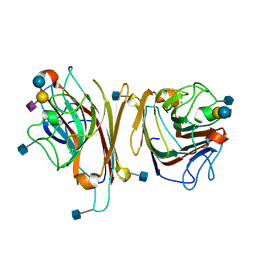 | | MAACKIA AMURENSIS LEUKOAGGLUTININ (LECTIN) WITH SIALYLLACTOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Imberty, A, Gautier, C, Lescar, J, Loris, R. | | Deposit date: | 1999-11-03 | | Release date: | 2000-06-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | An unusual carbohydrate binding site revealed by the structures of two Maackia amurensis lectins complexed with sialic acid-containing oligosaccharides.
J.Biol.Chem., 275, 2000
|
|
7E2O
 
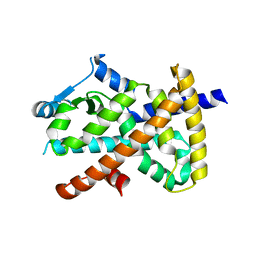 | | X-ray Crystal structure of PPARgamma R288H mutant. | | Descriptor: | Peroxisome proliferator-activated receptor gamma | | Authors: | Egawa, D, Itoh, T. | | Deposit date: | 2021-02-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insights into the Loss-of-Function R288H Mutant of Human PPAR gamma.
Biol.Pharm.Bull., 44, 2021
|
|
1WZE
 
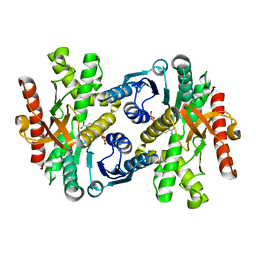 | |
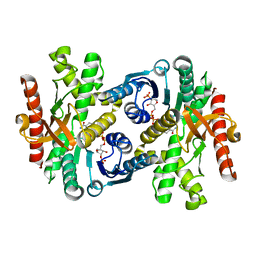
1WZI
 
 | |
1Y7T
 
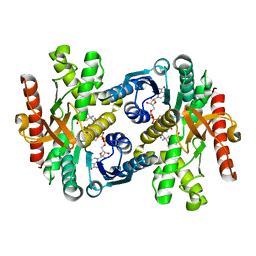 | | Crystal structure of NAD(H)-depenent malate dehydrogenase complexed with NADPH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Malate dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tomita, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2004-12-10 | | Release date: | 2005-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of NAD-dependent malate dehydrogenase complexed with NADP(H)
Biochem.Biophys.Res.Commun., 334, 2005
|
|
7C7V
 
 | | Vitamin D3 receptor/lithochoric acid derivative complex | | Descriptor: | (4R)-4-[(3R,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid, FORMIC ACID, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Masuno, H, Numoto, N, Kagechika, H, Ito, N. | | Deposit date: | 2020-05-26 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lithocholic Acid Derivatives as Potent Vitamin D Receptor Agonists.
J.Med.Chem., 64, 2021
|
|
7C7W
 
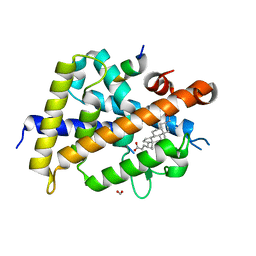 | | Vitamin D3 receptor/lithochoric acid derivative complex | | Descriptor: | (4R)-4-[(3S,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid, FORMIC ACID, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Masuno, H, Numoto, N, Kagechika, H, Ito, N. | | Deposit date: | 2020-05-26 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lithocholic Acid Derivatives as Potent Vitamin D Receptor Agonists.
J.Med.Chem., 64, 2021
|
|
6K5O
 
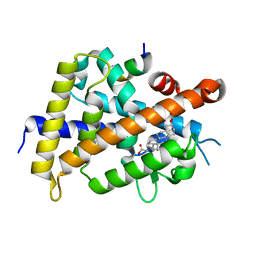 | | Development of Novel Lithocholic Acid Derivatives as Vitamin D Receptor Agonists | | Descriptor: | (4~{R})-4-[(3~{R},5~{R},8~{R},9~{S},10~{S},13~{R},14~{S},17~{R})-10,13-dimethyl-3-methylsulfonyloxy-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Masuno, H, Kagechika, H, Ito, N. | | Deposit date: | 2019-05-29 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of novel lithocholic acid derivatives as vitamin D receptor agonists.
Bioorg.Med.Chem., 27, 2019
|
|
2CVQ
 
 | | Crystal structure of NAD(H)-dependent malate dehydrogenase complexed with NADPH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Malate dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tomita, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2005-06-13 | | Release date: | 2005-08-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of NAD-dependent malate dehydrogenase complexed with NADP(H)
Biochem.Biophys.Res.Commun., 334, 2005
|
|
