5Z1N
 
 | | Crystal structure of C terminal region of G-protein interacting protein 1 (Gip1) from Dictyostelium discoideum | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, G-protein interacting protein 1, ... | | Authors: | Miyagawa, T, Koteishi, H, Kamimura, Y, Miyanaga, Y, Takeshita, K, Nakagawa, A, Ueda, M. | | Deposit date: | 2017-12-27 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural basis of Gip1 for cytosolic sequestration of G protein in wide-range chemotaxis
Nat Commun, 9, 2018
|
|
5Z39
 
 | | Crystal structure of C terminal region of G-protein interacting protein 1 (Gip1) from Dictyostelium discoideum form II | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, G-protein interacting protein 1, ... | | Authors: | Miyagawa, T, Koteishi, H, Kamimura, Y, Miyanaga, Y, Takeshita, K, Nakagawa, A, Ueda, M. | | Deposit date: | 2018-01-05 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural basis of Gip1 for cytosolic sequestration of G protein in wide-range chemotaxis
Nat Commun, 9, 2018
|
|
2EGY
 
 | | Crystal structure of LysN, alpha-aminoadipate aminotransferase (substrate free form), from Thermus thermophilus HB27 | | Descriptor: | Alpha-aminodipate aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tomita, T, Miyazaki, T, Miyagawa, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2007-03-02 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of LysN, alpha-aminoadipate aminotransferase, from Thermus thermophilus HB27
To be Published
|
|
2Z1Y
 
 | | Crystal structure of LysN, alpha-aminoadipate aminotransferase (complexed with N-(5'-phosphopyridoxyl)-L-leucine), from Thermus thermophilus HB27 | | Descriptor: | Alpha-aminodipate aminotransferase, LEUCINE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tomita, T, Miyazaki, T, Miyagawa, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2007-05-16 | | Release date: | 2008-06-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of broad substrate specificity of alpha-aminoadipate aminotransferase from Thermus thermophilus
To be Published
|
|
2ZP7
 
 | | Crystal structure of LysN, alpha-aminoadipate aminotransferase (Leucine complex), from Thermus thermophilus HB27 | | Descriptor: | Alpha-aminodipate aminotransferase, LEUCINE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tomita, T, Miyazaki, T, Miyagawa, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2008-06-30 | | Release date: | 2009-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Mechanism for multiple-substrates recognition of alpha-aminoadipate aminotransferase from Thermus thermophilus
Proteins, 2008
|
|
3CBF
 
 | | Crystal structure of LysN, alpha-aminoadipate aminotransferase, from Thermus thermophilus HB27 | | Descriptor: | (2S)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]hexanedioic acid, Alpha-aminodipate aminotransferase | | Authors: | Tomita, T, Miyazaki, T, Miyagawa, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2008-02-21 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Mechanism for multiple-substrates recognition of alpha-aminoadipate aminotransferase from Thermus thermophilus
Proteins, 2008
|
|
1C53
 
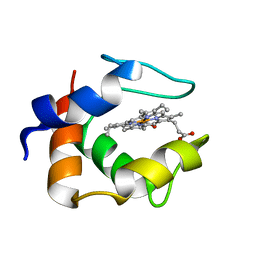 | | S-CLASS CYTOCHROMES C HAVE A VARIETY OF FOLDING PATTERNS: STRUCTURE OF CYTOCHROME C-553 FROM DESULFOVIBRIO VULGARIS DETERMINED BY THE MULTI-WAVELENGTH ANOMALOUS DISPERSION METHOD | | Descriptor: | CYTOCHROME C553, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nakagawa, A, Higuchi, Y, Yasuoka, N, Katsube, Y, Yaga, T. | | Deposit date: | 1991-08-26 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | S-class cytochromes c have a variety of folding patterns: structure of cytochrome c-553 from Desulfovibrio vulgaris determined by the multi-wavelength anomalous dispersion method.
J.Biochem.(Tokyo), 108, 1990
|
|
4NWL
 
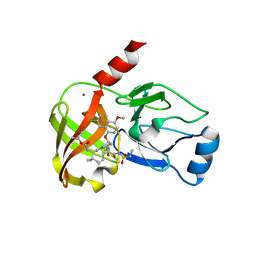 | | Crystal structure of hepatis c virus protease (ns3) complexed with bms-650032 aka n-(tert-butoxycarbonyl)-3-me thyl-l-valyl-(4r)-4-((7-chloro-4-methoxy-1-isoquinolinyl)o xy)-n-((1r,2s)-1-((cyclopropylsulfonyl)carbamoyl)-2-vinylc yclopropyl)-l-prolinamide | | Descriptor: | HCV NS3 1a Protease, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-4-[(7-chloro-4-methoxyisoquinolin-1-yl)oxy]-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-L-prolinamide, ZINC ION | | Authors: | Muckelbauer, J.K, Klei, H.E. | | Deposit date: | 2013-12-06 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Early Clinical Evaluation of BMS-605339, a Potent and Orally Efficacious Tripeptidic Acylsulfonamide NS3 Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 57, 2014
|
|
4NWK
 
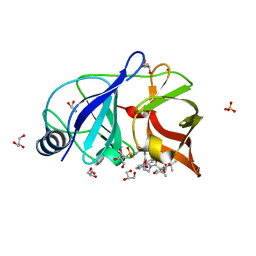 | | Crystal structure of hepatis c virus protease (ns3) complexed with bms-605339 aka n-(tert-butoxycarbonyl)-3-me thyl-l-valyl-(4r)-n-((1r,2s)-1-((cyclopropylsulfonyl)carba moyl)-2-vinylcyclopropyl)-4-((6-methoxy-1-isoquinolinyl)ox y)-l-prolinamide | | Descriptor: | GLYCEROL, HCV NS3 1a Protease, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-4-[(6-methoxyisoquinolin-1-yl)oxy]-L-prolinamide, ... | | Authors: | Muckelbauer, J.K, Klei, H.E. | | Deposit date: | 2013-12-06 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Early Clinical Evaluation of BMS-605339, a Potent and Orally Efficacious Tripeptidic Acylsulfonamide NS3 Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 57, 2014
|
|
7MCF
 
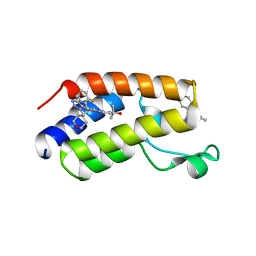 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH 2-{3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-6-fluoro-5-[(S)-(3-fluoropyridin-2-yl)(oxan-4-yl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol | | Descriptor: | 2-{3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-6-fluoro-5-[(S)-(3-fluoropyridin-2-yl)(oxan-4-yl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol, Bromodomain-containing protein 4 | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-02 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Development of BET inhibitors as potential treatments for cancer: A search for structural diversity.
Bioorg.Med.Chem.Lett., 44, 2021
|
|
7MCE
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH 2-{(7P)-7-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-8-fluoro-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-3-yl}propan-2-ol | | Descriptor: | 2-{(7P)-7-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-8-fluoro-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-3-yl}propan-2-ol, Bromodomain-containing protein 4 | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-02 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Development of BET inhibitors as potential treatments for cancer: A new carboline chemotype.
Bioorg.Med.Chem.Lett., 51, 2021
|
|
2ZYJ
 
 | | Crystal structure of LysN, alpha-aminoadipate aminotransferase (complexed with N-(5'-phosphopyridoxyl)-L-glutamate), from Thermus thermophilus HB27 | | Descriptor: | Alpha-aminodipate aminotransferase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-glutamic acid | | Authors: | Ouchi, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2009-01-26 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Dual roles of a conserved pair, Arg23 and Ser20, in recognition of multiple substrates in alpha-aminoadipate aminotransferase from Thermus thermophilus.
Biochem.Biophys.Res.Commun., 388, 2009
|
|
6XP9
 
 | |
7UZN
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH BMT-206059 AKA 2-{(3M)-3-(1,4-DIMETHYL-1H-1,2,3-TRIAZOL-5-YL)-8-FLUORO-5-[(S)-(OXAN-4-YL)(PHENYL)METHYL]-5H-PYRIDO[3,2-b]INDOL-7-YL}PROPAN-2-OL, TRIPLY DEUTERATED ON THE 4-METHYL GROUP | | Descriptor: | 1,2-ETHANEDIOL, 2-{(3M)-3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-8-fluoro-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol, Bromodomain-containing protein 4, ... | | Authors: | Sheriff, S. | | Deposit date: | 2022-05-09 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.685 Å) | | Cite: | Development of BET Inhibitors as Potential Treatments for Cancer: Optimization of Pharmacokinetic Properties.
Acs Med.Chem.Lett., 13, 2022
|
|
5TWM
 
 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS GENOTYPE 2A STRAIN JFH1 L30S NS5B RNA-DEPENDENT RNA POLYMERASE IN COMPLEX WITH 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide, NS5B RNA-dependent RNA POLYMERASE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The discovery of a pan-genotypic, primer grip inhibitor of HCV NS5B polymerase.
Medchemcomm, 8, 2017
|
|
5TWN
 
 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA- DEPENDENT RNA POLYMERASE IN COMPLEX WITH 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide, NS5B RNA- DEPENDENT RNA POLYMERASE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | The discovery of a pan-genotypic, primer grip inhibitor of HCV NS5B polymerase.
Medchemcomm, 8, 2017
|
|
