8QC3
 
 | |
8QC2
 
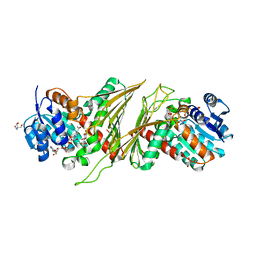 | | Crystal structure of NAD-dependent glycoside hydrolase from Flavobacterium sp. (strain K172) in complex with co-factor NAD+ and sulfoquinovose (SQ) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Pickles, I.B, Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Widespread Family of NAD + -Dependent Sulfoquinovosidases at the Gateway to Sulfoquinovose Catabolism.
J.Am.Chem.Soc., 145, 2023
|
|
5N0F
 
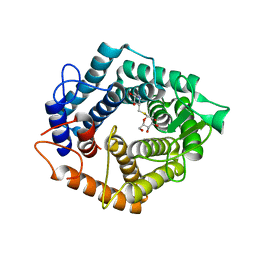 | | The catalytic domain, BcGH76, of Bacillus circulans Aman6 in complex with 1,6-ManSIFG | | Descriptor: | Alpha-1,6-mannanase, [(3S,4R,5R)-4,5-dihydroxypiperidin-3-yl]methyl 1-thio-alpha-D-mannopyranoside | | Authors: | Jin, Y, Williams, S, Davies, G. | | Deposit date: | 2017-02-02 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | An atypical interaction explains the high-affinity of a non-hydrolyzable S-linked 1,6-alpha-mannanase inhibitor.
Chem. Commun. (Camb.), 53, 2017
|
|
3AXL
 
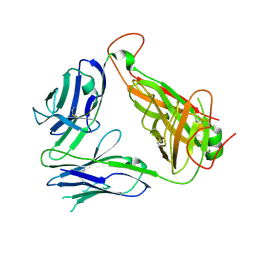 | | Murine Valpha 10 Vbeta 8.1 T-cell receptor | | Descriptor: | Valpha 10, Vbeta 8.1 | | Authors: | Patel, O, Rossjohn, J. | | Deposit date: | 2011-04-11 | | Release date: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A semi-invariant V(alpha)10(+) T cell antigen receptor defines a population of natural killer T cells with distinct glycolipid antigen-recognition properties
Nat.Immunol., 12, 2011
|
|
7AGH
 
 | |
7AG7
 
 | |
7AG4
 
 | |
7AG6
 
 | |
7AG1
 
 | |
7AGK
 
 | | Crystal structure of E. coli SF kinase (YihV) in complex with product sulfofructose phosphate (SFP) | | Descriptor: | Sulfofructose kinase, [(2~{S},3~{S},4~{S},5~{R})-3,4,5-tris(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]methanesulfonic acid | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Molecular Basis of Sulfosugar Selectivity in Sulfoglycolysis.
Acs Cent.Sci., 7, 2021
|
|
7BC0
 
 | |
7BBY
 
 | |
7BC1
 
 | |
7BBZ
 
 | |
5TEB
 
 | | Crystal Structure of the TIR domain from the Arabidopsis Thaliana disease resistance protein RPP1 | | Descriptor: | Recognition of Peronospora parasitica 1 | | Authors: | Bentham, A.R, Zhang, X, Croll, T, Williams, S, Kobe, B. | | Deposit date: | 2016-09-20 | | Release date: | 2017-02-01 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Multiple functional self-association interfaces in plant TIR domains.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6MSS
 
 | | Diversity in the type II Natural Killer T cell receptor repertoire and antigen specificity leads to differing CD1d docking strategies | | Descriptor: | (2S)-2-(heptadecanoyloxy)-3-{[(10S)-10-methyloctadecanoyl]oxy}propyl alpha-D-glucopyranosiduronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sundararaj, S, Le Nours, J, Praveena, T, Rossjohn, J. | | Deposit date: | 2018-10-18 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Distinct CD1d docking strategies exhibited by diverse Type II NKT cell receptors.
Nat Commun, 10, 2019
|
|
6MRA
 
 | | Diversity in the type II Natural Killer T cell receptor repertoire and antigen specificity leads to differing CD1d docking strategies | | Descriptor: | TCR alpha chain, TCR beta-chain | | Authors: | Sundararaj, S, Le Nours, J, Praveena, T, Rossjohn, J. | | Deposit date: | 2018-10-12 | | Release date: | 2019-09-18 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Distinct CD1d docking strategies exhibited by diverse Type II NKT cell receptors.
Nat Commun, 10, 2019
|
|
6RQK
 
 | | Crystal structure of GH125 1,6-alpha-mannosidase from Clostridium perfringens in complex with mannoimidazole | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, Alpha-1,6-mannosidase | | Authors: | Males, A, Davies, G.J. | | Deposit date: | 2019-05-16 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Distortion of mannoimidazole supports a B2,5boat transition state for the family GH125 alpha-1,6-mannosidase from Clostridium perfringens.
Org.Biomol.Chem., 17, 2019
|
|
6SMY
 
 | |
8AH3
 
 | |
8BC2
 
 | | Ligand-Free Structure of the decameric sulfofructose transaldolase BmSF-TAL | | Descriptor: | Transaldolase | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
8BC4
 
 | | Cryo-EM Structure of a BmSF-TAL - Sulfofructose Schiff Base Complex in symmetry group C1 | | Descriptor: | (2~{R},3~{S},4~{S})-2,3,4,6-tetrakis(oxidanyl)hexane-1-sulfonic acid, Transaldolase | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
8BC3
 
 | | Cryo-EM Structure of a BmSF-TAL - Sulfofructose Schiff Base Complex | | Descriptor: | (2~{R},3~{S},4~{S})-2,3,4,6-tetrakis(oxidanyl)hexane-1-sulfonic acid, BmSF-TAL | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
8C4I
 
 | |
6SMZ
 
 | |
