7JZL
 
 | |
7JZN
 
 | |
7JZM
 
 | |
7JZU
 
 | |
7KI6
 
 | |
7KI4
 
 | |
7M51
 
 | | B6 Fab fragment bound to the OC43 spike stem helix peptide | | Descriptor: | B6 antigen-binding (Fab) fragment heavy chain, B6 antigen-binding (Fab) fragment light chain, GLYCEROL, ... | | Authors: | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M55
 
 | | B6 Fab fragment bound to the MERS-CoV spike stem helix peptide | | Descriptor: | B6 antigen binding fragment (Fab) heavy chain, B6 antigen binding fragment (Fab) light chain, GLYCEROL, ... | | Authors: | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M5E
 
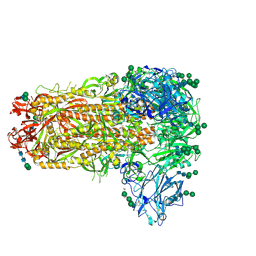 | |
7M53
 
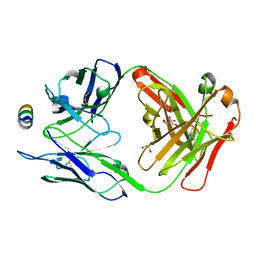 | | B6 Fab fragment bound to the SARS-CoV/SARS-CoV-2 spike stem helix peptide | | Descriptor: | B6 antigen-binding (Fab) fragment heavy chain, B6 antigen-binding (Fab) fragment light chain, GLYCEROL, ... | | Authors: | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M52
 
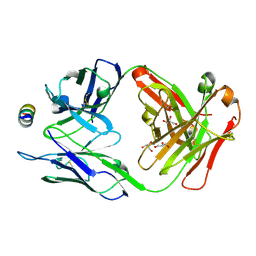 | | B6 Fab fragment bound to the HKU4 spike stem helix peptide | | Descriptor: | B6 antigen-binding (Fab) fragment heavy chain, B6 antigen-binding (Fab) fragment light chain, GLYCEROL, ... | | Authors: | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4IL7
 
 | |
5HX2
 
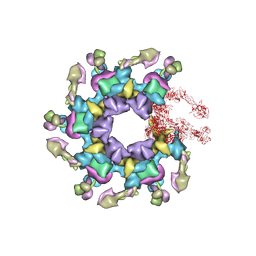 | | In vitro assembled star-shaped hubless T4 baseplate | | Descriptor: | Baseplate wedge protein gp10, Baseplate wedge protein gp53, Baseplate wedge protein gp6, ... | | Authors: | Yap, M.L, Klose, T, Fokine, A, Rossmann, M.G. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Role of bacteriophage T4 baseplate in regulating assembly and infection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6XSS
 
 | | CryoEM structure of designed helical fusion protein C4_nat_HFuse-7900 | | Descriptor: | C4_nat_HFuse-7900 | | Authors: | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2020-07-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
6XNS
 
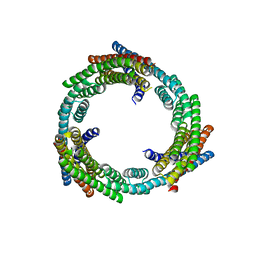 | | C3_crown-05 | | Descriptor: | C3_crown-05 | | Authors: | Bick, M.J, Hsia, Y, Sankaran, B, Baker, D. | | Deposit date: | 2020-07-04 | | Release date: | 2020-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
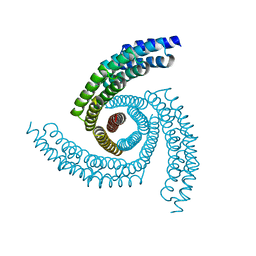
6XT4
 
 | |
6XI6
 
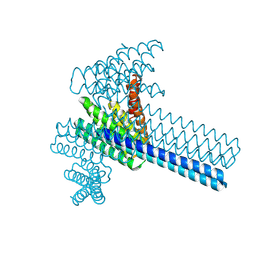 | | Hierarchical design of multi-scale protein complexes by combinatorial assembly of oligomeric helical bundle and repeat protein building blocks | | Descriptor: | helical fusion design | | Authors: | Bera, A.K, Hsia, Y, Kang, A.S, Shankaran, B, Baker, D. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
6XH5
 
 | |
7R6X
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2E12 Fab, S309 Fab, and S304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Monoclonal antibody S2E12 Fab heavy chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
3EJC
 
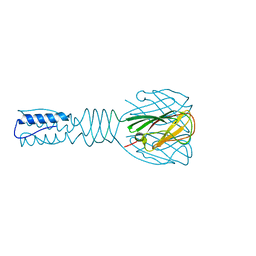 | | Full length Receptor Binding Protein from Lactococcal phage TP901-1 | | Descriptor: | Baseplate protein (BPP) | | Authors: | Spinelli, S, Lichiere, J, Blangy, S, Sciara, G, Cambillau, C, Campanacci, V. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and molecular assignment of lactococcal phage TP901-1 baseplate.
J.Biol.Chem., 285, 2010
|
|
6DRH
 
 | | ADP-ribosyltransferase toxin/immunity pair | | Descriptor: | ADP-ribosyl-(Dinitrogen reductase) hydrolase, MAGNESIUM ION, PAAR repeat-containing protein, ... | | Authors: | Bosch, D.E, Ting, S, Allaire, M, Mougous, J.D. | | Deposit date: | 2018-06-11 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Bifunctional Immunity Proteins Protect Bacteria against FtsZ-Targeting ADP-Ribosylating Toxins.
Cell, 175, 2018
|
|
6DRE
 
 | | ADP-ribosyltransferase toxin/immunity pair | | Descriptor: | ADP-ribosyl-(Dinitrogen reductase) hydrolase, MAGNESIUM ION, PAAR repeat-containing protein | | Authors: | Bosch, D.E, Ting, S, Allaire, M, Mougous, J.D. | | Deposit date: | 2018-06-11 | | Release date: | 2018-10-31 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bifunctional Immunity Proteins Protect Bacteria against FtsZ-Targeting ADP-Ribosylating Toxins.
Cell, 175, 2018
|
|
6V8E
 
 | |
6VEH
 
 | |
6VFJ
 
 | |
