7F8K
 
 | |
1AV4
 
 | |
1AVL
 
 | |
7WNO
 
 | |
7WNP
 
 | |
7YNH
 
 | |
2GLT
 
 | | STRUCTURE OF ESCHERICHIA COLI GLUTATHIONE SYNTHETASE AT PH 6.0. | | Descriptor: | GLUTATHIONE BIOSYNTHETIC LIGASE | | Authors: | Matsuda, K, Yamaguchi, H, Kato, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-05-16 | | Release date: | 1995-07-31 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glutathione synthetase at optimal pH: domain architecture and structural similarity with other proteins.
Protein Eng., 9, 1996
|
|
1BD0
 
 | | ALANINE RACEMASE COMPLEXED WITH ALANINE PHOSPHONATE | | Descriptor: | ALANINE RACEMASE, {1-[(3-HYDROXY-METHYL-5-PHOSPHONOOXY-METHYL-PYRIDIN-4-YLMETHYL)-AMINO]-ETHYL}-PHOSPHONIC ACID | | Authors: | Stamper, G.F, Morollo, A.A, Ringe, D. | | Deposit date: | 1998-05-12 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Reaction of alanine racemase with 1-aminoethylphosphonic acid forms a stable external aldimine.
Biochemistry, 37, 1998
|
|
1SFT
 
 | | ALANINE RACEMASE | | Descriptor: | ACETATE ION, ALANINE RACEMASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shaw, J.P, Petsko, G.A, Ringe, D. | | Deposit date: | 1996-09-20 | | Release date: | 1997-02-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of the structure of alanine racemase from Bacillus stearothermophilus at 1.9-A resolution.
Biochemistry, 36, 1997
|
|
2SFP
 
 | | ALANINE RACEMASE WITH BOUND PROPIONATE INHIBITOR | | Descriptor: | PROPANOIC ACID, PROTEIN (ALANINE RACEMASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Morollo, A.A, Petsko, G.A, Ringe, D. | | Deposit date: | 1999-02-16 | | Release date: | 1999-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Michaelis complex analogue: propionate binds in the substrate carboxylate site of alanine racemase.
Biochemistry, 38, 1999
|
|
1JMX
 
 | |
1JMZ
 
 | | crystal structure of a quinohemoprotein amine dehydrogenase from pseudomonas putida with inhibitor | | Descriptor: | Amine Dehydrogenase, HEME C, NICKEL (II) ION, ... | | Authors: | Satoh, A, Miyahara, I, Hirotsu, K. | | Deposit date: | 2001-07-20 | | Release date: | 2002-01-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of quinohemoprotein amine dehydrogenase from Pseudomonas putida. Identification of a novel quinone cofactor encaged by multiple thioether cross-bridges.
J.Biol.Chem., 277, 2002
|
|
2ZL8
 
 | |
2ZWM
 
 | |
2ZXJ
 
 | |
1GSH
 
 | | STRUCTURE OF ESCHERICHIA COLI GLUTATHIONE SYNTHETASE AT PH 7.5 | | Descriptor: | GLUTATHIONE BIOSYNTHETIC LIGASE | | Authors: | Matsuda, K, Kato, H, Yamaguchi, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-05-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glutathione synthetase at optimal pH: domain architecture and structural similarity with other proteins.
Protein Eng., 9, 1996
|
|
2D0W
 
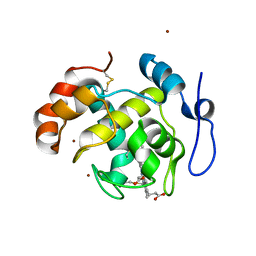 | | Crystal structure of cytochrome cL from Hyphomicrobium denitrificans | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, ZINC ION, cytochrome cL | | Authors: | Nojiri, M, Hira, D, Yamaguchi, K, Suzuki, S. | | Deposit date: | 2005-08-10 | | Release date: | 2006-08-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of cytochrome c(L) and methanol dehydrogenase from Hyphomicrobium denitrificans: structural and mechanistic insights into interactions between the two proteins
Biochemistry, 45, 2006
|
|
2D0V
 
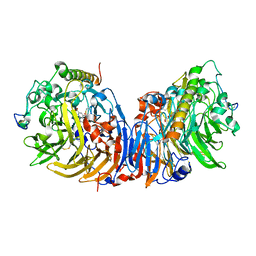 | | Crystal structure of methanol dehydrogenase from Hyphomicrobium denitrificans | | Descriptor: | CALCIUM ION, PYRROLOQUINOLINE QUINONE, methanol dehydrogenase large subunit, ... | | Authors: | Nojiri, M, Hira, D, Yamaguchi, K, Suzuki, S. | | Deposit date: | 2005-08-09 | | Release date: | 2006-08-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structures of cytochrome c(L) and methanol dehydrogenase from Hyphomicrobium denitrificans: structural and mechanistic insights into interactions between the two proteins
Biochemistry, 45, 2006
|
|
