1IU8
 
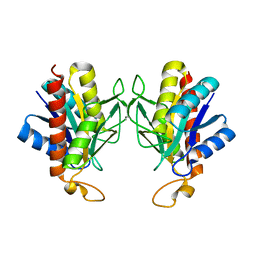 | | The X-ray Crystal Structure of Pyrrolidone-Carboxylate Peptidase from Hyperthermophilic Archaeon Pyrococcus horikoshii | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Sokabe, M, Kawamura, T, Sakai, N, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2002-02-28 | | Release date: | 2002-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The X-ray crystal structure of pyrrolidone-carboxylate peptidase from hyperthermophilic archaea Pyrococcus horikoshii
J.STRUCT.FUNCT.GENOM., 2, 2002
|
|
1ION
 
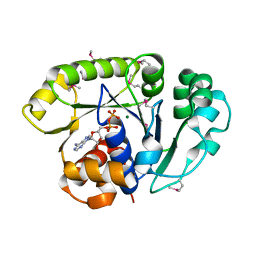 | | THE SEPTUM SITE-DETERMINING PROTEIN MIND COMPLEXED WITH MG-ADP FROM PYROCOCCUS HORIKOSHII OT3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROBABLE CELL DIVISION INHIBITOR MIND | | Authors: | Sakai, N, Yao, M, Itou, H, Watanabe, N, Yumoto, F, Tanokura, M, Tanaka, I. | | Deposit date: | 2001-03-21 | | Release date: | 2001-09-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of septum site-determining protein MinD from Pyrococcus horikoshii OT3 in complex with Mg-ADP.
Structure, 9, 2001
|
|
1IQV
 
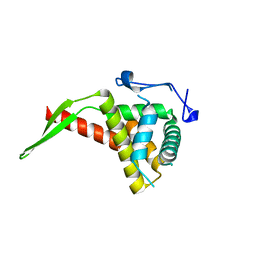 | |
1VCX
 
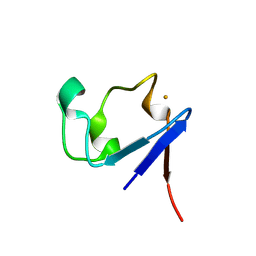 | | Neutron Crystal Structure of the Wild Type Rubredoxin from Pyrococcus Furiosus at 1.5A Resolution | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Kurihara, K, Tanaka, I, Chatake, T, Adams, M.W.W, Jenney Jr, F.E, Moiseeva, N, Bau, R, Niimura, N. | | Deposit date: | 2004-03-17 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.5 Å) | | Cite: | Neutron crystallographic study on rubredoxin from Pyrococcus furiosus by BIX-3, a single-crystal diffractometer for biomacromolecules
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1VGJ
 
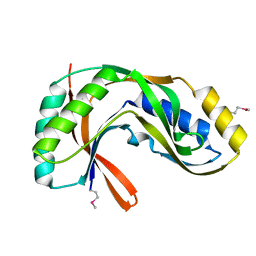 | | Crystal structure of 2'-5' RNA ligase from Pyrococcus horikoshii | | Descriptor: | Hypothetical protein PH0099 | | Authors: | Yao, M, Morita, H, Okada, A, Tanaka, I. | | Deposit date: | 2004-04-27 | | Release date: | 2005-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The structure of Pyrococcus horikoshii 2'-5' RNA ligase at 1.94 A resolution reveals a possible open form with a wider active-site cleft
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
1VCI
 
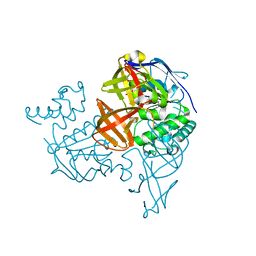 | | Crystal structure of the ATP-binding cassette of multisugar transporter from Pyrococcus horikoshii OT3 complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, sugar-binding transport ATP-binding protein | | Authors: | Ose, T, Fujie, T, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2004-03-08 | | Release date: | 2004-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the ATP-binding cassette of multisugar transporter from Pyrococcus horikoshii OT3
Proteins, 57, 2004
|
|
1VDZ
 
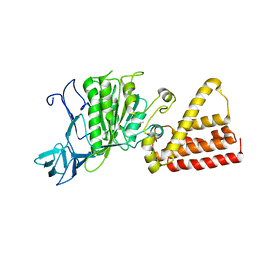 | | Crystal structure of A-type ATPase catalytic subunit A from Pyrococcus horikoshii OT3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, A-type ATPase subunit A | | Authors: | Maegawa, Y, Morita, H, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2004-03-26 | | Release date: | 2005-06-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of A-type ATPase catalytic subunit A from Pyrococcus horikoshii OT3
To be Published
|
|
1VGG
 
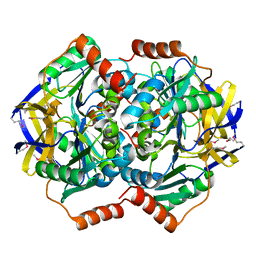 | | Crystal Structure of the Conserved Hypothetical Protein TTHA1091 from Thermus Thermophilus HB8 | | Descriptor: | Conserved Hypothetical Protein TT1634 (TTHA1091) | | Authors: | Satoh, S, Yao, M, Kousumi, Y, Ebihara, A, Matsumoto, K, Okamoto, A, Tanaka, I, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-04-26 | | Release date: | 2004-10-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Conserved Hypothetical Protein TT1634 from Thermus Thermophilus HB8
To be Published
|
|
1WLT
 
 | | Crystal Structure of dTDP-4-dehydrorhamnose 3,5-epimerase homologue from Sulfolobus tokodaii | | Descriptor: | 176aa long hypothetical dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Watanabe, N, Sakai, N, Zilian, Z, Kawarabayasi, Y, Tanaka, I. | | Deposit date: | 2004-06-29 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of dTDP-4-dehydrorhamnose 3,5-epimerase homologue
to be published
|
|
1WMI
 
 | | Crystal structure of archaeal RelE-RelB complex from Pyrococcus horikoshii OT3 | | Descriptor: | hypothetical protein PHS013, hypothetical protein PHS014 | | Authors: | Takagi, H, Kakuta, Y, Kamachi, R, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2004-07-09 | | Release date: | 2005-03-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of archaeal toxin-antitoxin RelE-RelB complex with implications for toxin activity and antitoxin effects
Nat.Struct.Mol.Biol., 12, 2005
|
|
1WQA
 
 | |
1WNU
 
 | | Structure of Archaeal Trans-Editing Protein AlaX in complex with L-serine | | Descriptor: | SERINE, ZINC ION, alanyl-tRNA synthetase | | Authors: | Sokabe, M, Okada, A, Nakashima, T, Yao, M, Tanaka, I. | | Deposit date: | 2004-08-09 | | Release date: | 2005-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of alanine discrimination in editing site
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1WY7
 
 | |
1WOE
 
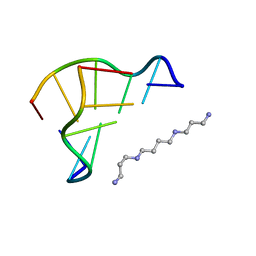 | | X-ray structure of a Z-DNA hexamer d(CGCGCG) | | Descriptor: | SPERMINE, Z-DNA hexamer | | Authors: | Chatake, T, Tanaka, I, Umino, H, Arai, S, Niimura, N. | | Deposit date: | 2004-08-15 | | Release date: | 2005-07-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The hydration structure of a Z-DNA hexameric duplex determined by a neutron diffraction technique.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7VEI
 
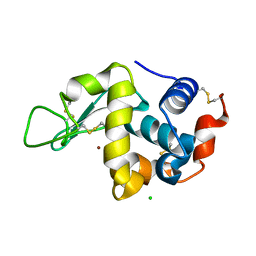 | | Neutron structure of D2O-solvent lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, NICKEL (II) ION | | Authors: | Chatake, T, Tanaka, I, Kusaka, K, Fujiwara, S. | | Deposit date: | 2021-09-08 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | Protonation states of hen egg-white lysozyme observed using D/H contrast neutron crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1WE5
 
 | | Crystal Structure of Alpha-Xylosidase from Escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative family 31 glucosidase yicI | | Authors: | Ose, T, Kitamura, M, Okuyama, M, Mori, H, Kimura, A, Watanabe, N, Yao, M, Tanaka, I. | | Deposit date: | 2004-05-24 | | Release date: | 2005-02-15 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Alpha-Xylosidase from Escherichia coli
TO BE PUBLISHED
|
|
1WLS
 
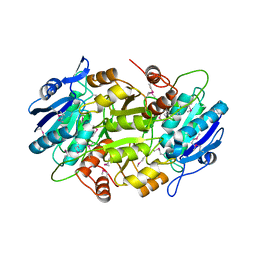 | | Crystal structure of L-asparaginase I homologue protein from Pyrococcus horikoshii | | Descriptor: | L-asparaginase | | Authors: | Yao, M, Morita, H, Yasutake, Y, Tanaka, I. | | Deposit date: | 2004-06-29 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of the type I L-asparaginase from the hyperthermophilic archaeon Pyrococcus horikoshii at 2.16 angstroms resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1WXO
 
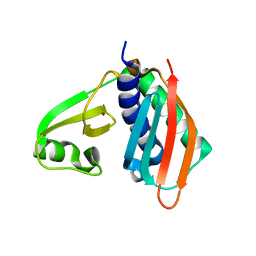 | | Structure of Archaeal Trans-Editing Protein AlaX in complex with zinc | | Descriptor: | ZINC ION, alanyl-tRNA synthetase | | Authors: | Sokabe, M, Okada, A, Nakashima, T, Yao, M, Tanaka, I. | | Deposit date: | 2005-01-27 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Molecular basis of alanine discrimination in editing site
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1WQZ
 
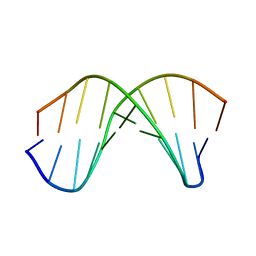 | | Complicated water orientations in the minor groove of B-DNA decamer D(CCATTAATGG)2 observed by neutron diffraction measurements | | Descriptor: | 5'-D(*CP*CP*AP*TP*TP*AP*AP*TP*GP*G)-3' | | Authors: | Arai, S, Chatake, T, Ohhara, T, Kurihara, K, Tanaka, I, Suzuki, N, Fujimoto, Z, Mizuno, H, Niimura, N. | | Deposit date: | 2004-10-07 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (3 Å) | | Cite: | Complicated water orientations in the minor groove of the B-DNA decamer d(CCATTAATGG)2 observed by neutron diffraction measurements
Nucleic Acids Res., 33, 2005
|
|
1WQY
 
 | | X-RAY structural analysis of B-DNA decamer D(CCATTAATGG)2 crystal grown in D2O solution | | Descriptor: | 5'-D(*CP*CP*AP*TP*TP*AP*AP*TP*GP*G)-3' | | Authors: | Arai, S, Chatake, T, Ohhara, T, Kurihara, K, Tanaka, I, Suzuki, N, Fujimoto, Z, Mizuno, H, Niimura, N. | | Deposit date: | 2004-10-07 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complicated water orientations in the minor groove of the B-DNA decamer d(CCATTAATGG)2 observed by neutron diffraction measurements
Nucleic Acids Res., 33, 2005
|
|
1WZC
 
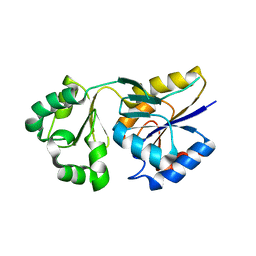 | | Crystal structure of Pyrococcus horikoshii mannosyl-3-phosphoglycerate phosphatase complexed with MG2+ and phosphate | | Descriptor: | MAGNESIUM ION, Mannosyl-3-phosphoglycerate phosphatase, PHOSPHATE ION | | Authors: | Kawamura, T, Watanabe, N, Tanaka, I. | | Deposit date: | 2005-03-04 | | Release date: | 2006-02-28 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of mannosyl-3-phosphoglycerate phosphatase from Pyrococcus horikoshii.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1WZZ
 
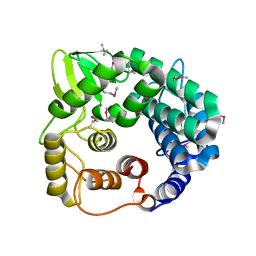 | | Structure of endo-beta-1,4-glucanase CMCax from Acetobacter xylinum | | Descriptor: | Probable endoglucanase, SULFATE ION | | Authors: | Yasutake, Y, Kawano, S, Tajima, K, Yao, M, Satoh, Y, Munekata, M, Tanaka, I, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-10 | | Release date: | 2006-03-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural characterization of the Acetobacter xylinum endo-beta-1,4-glucanase CMCax required for cellulose biosynthesis.
Proteins, 64, 2006
|
|
3VZR
 
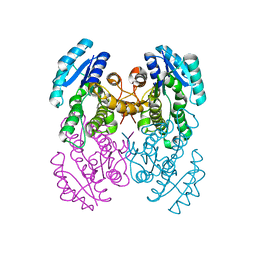 | |
3VZQ
 
 | |
3VSE
 
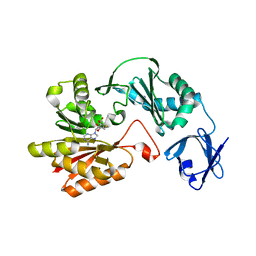 | | Crystal structure of methyltransferase | | Descriptor: | Putative uncharacterized protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kita, S, Tanaka, Y, Yao, M, Tanaka, I. | | Deposit date: | 2012-04-25 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Crystal structure of a putative methyltransferase SAV1081 from Staphylococcus aureus
Protein Pept.Lett., 20, 2012
|
|
