2HS5
 
 | | Structural Genomics, the crystal structure of a putative transcriptional regulator GntR from Rhodococcus sp. RHA1 | | Descriptor: | ACETATE ION, putative transcriptional regulator GntR | | Authors: | Tan, K, Skarina, T, Onopriyenko, A, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-21 | | Release date: | 2006-08-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a putative transcriptional regulator GntR from Rhodococcus sp. RHA1
To be Published
|
|
2HKU
 
 | | Structural Genomics, the crystal structure of a putative transcriptional regulator from Rhodococcus sp. RHA1 | | Descriptor: | 1,2-ETHANEDIOL, A putative transcriptional regulator, TETRAETHYLENE GLYCOL | | Authors: | Tan, K, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a transcriptional regulator from Rhodococcus sp. RHA1
To be Published
|
|
2I71
 
 | | Crystal structure of a Conserved Protein of Unknown Function from Sulfolobus solfataricus P2 | | Descriptor: | Hypothetical protein, MAGNESIUM ION | | Authors: | Tan, K, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a Conserved hypothetical protein from Sulfolobus solfataricus P2
To be Published
|
|
2HXI
 
 | | Structural Genomics, the crystal structure of a putative transcriptional regulator from Streptomyces coelicolor A3(2) | | Descriptor: | Putative transcriptional regulator | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-03 | | Release date: | 2006-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a putative transcriptional regulator TetR from Streptomyces coelicolor A3(2)
To be Published
|
|
2I5E
 
 | | Crystal Structure of a Protein of Unknown Function MM2497 from Methanosarcina mazei Go1, Probable Nucleotidyltransferase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Hypothetical protein MM_2497 | | Authors: | Tan, K, Du, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-24 | | Release date: | 2006-09-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a hypothetical protein MM_2497 from Methanosarcina mazei Go1
To be Published
|
|
2I9X
 
 | | Structural Genomics, the crystal structure of SpoVG conserved domain from Staphylococcus epidermidis ATCC 12228 | | Descriptor: | 1,2-ETHANEDIOL, Putative septation protein spoVG | | Authors: | Tan, K, Maltseva, N, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-06 | | Release date: | 2006-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of SpoVG from Staphylococcus epidermidis ATCC 12228
To be Published
|
|
2IA9
 
 | | Structural Genomics, the crystal structure of SpoVG from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative septation protein spoVG, SULFATE ION | | Authors: | Tan, K, Borovilos, M, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-07 | | Release date: | 2006-10-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of SpoVG from Bacillus subtilis subsp. subtilis str. 168
To be Published
|
|
2I9Z
 
 | | Structural Genomics, the Crystal structure of full-length SpoVG from Staphylococcus epidermidis ATCC 12228 | | Descriptor: | 1,2-ETHANEDIOL, Putative septation protein spoVG | | Authors: | Tan, K, Maltseva, N, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-06 | | Release date: | 2006-10-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal structure of SpoVG from Staphylococcus epidermidis ATCC 12228
To be Published
|
|
2IKK
 
 | | Structural Genomics, the crystal structure of the C-terminal domain of Yurk from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | Hypothetical transcriptional regulator yurK, SULFATE ION | | Authors: | Tan, K, Hatzos, C, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the C-terminal domain of Yurk from Bacillus subtilis subsp. subtilis str. 168
To be Published
|
|
5CJJ
 
 | | The crystal structure of phosphoribosylglycinamide formyltransferase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-07-14 | | Release date: | 2015-07-29 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The crystal structure of phosphoribosylglycinamide formyltransferase from Campylobacter jejuni subsp. jejuni NCTC 11168
To Be Published
|
|
5EUF
 
 | | The crystal structure of a protease from Helicobacter pylori | | Descriptor: | GLYCEROL, Protease, ZINC ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-18 | | Release date: | 2015-12-02 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of a protease from Helicobacter pylori
To Be Published
|
|
5EWQ
 
 | | The crystal structure of an amidase family protein from Bacillus anthracis str. Ames | | Descriptor: | ACETATE ION, Amidase | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-20 | | Release date: | 2015-12-09 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The crystal structure of an amidase family protein from Bacillus anthracis str. Ames
To Be Published
|
|
7KB7
 
 | | THE STRUCTURE OF A SENSOR DOMAIN OF A HISTIDINE KINASE (VxrA) FROM VIBRIO CHOLERAE O1 BIOVAR ELTOR STR. N16961, N239-T240 deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Tan, K, Wu, R, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Membrane Proteins of Infectious Diseases (MPID) | | Deposit date: | 2020-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 2021
|
|
7KB3
 
 | | The structure of a sensor domain of a histidine kinase (VxrA) from Vibrio cholerae O1 biovar eltor str. N16961, 2nd form | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, K, Wu, R, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 2021
|
|
7KB9
 
 | | THE STRUCTURE OF A SENSOR DOMAIN OF A HISTIDINE KINASE (VxrA) FROM VIBRIO CHOLERAE O1 BIOVAR ELTOR STR. N16961, D238-T240 deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Sensor histidine kinase | | Authors: | Tan, K, Wu, R, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 2021
|
|
7LA6
 
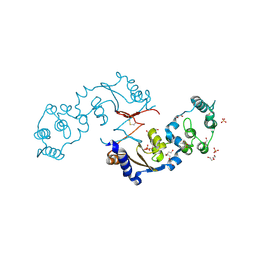 | | THE STRUCTURE OF A SENSOR DOMAIN OF A HISTIDINE KINASE (VxrA) FROM VIBRIO CHOLERAE O1 BIOVAR ELTOR STR. N16961, N239 deletion mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Wu, R, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-05 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 203, 2021
|
|
3IKB
 
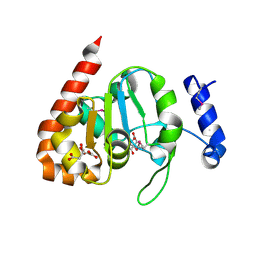 | |
3ILK
 
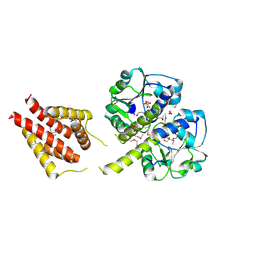 | | The structure of a probable methylase family protein from Haemophilus influenzae Rd KW20 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Tan, K, Li, H, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-07 | | Release date: | 2009-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The structure of a probable methylase family protein from Haemophilus influenzae Rd KW20
To be Published
|
|
3KKB
 
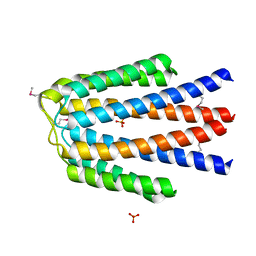 | |
3KBR
 
 | | The crystal structure of cyclohexadienyl dehydratase precursor from Pseudomonas aeruginosa PA01 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Cyclohexadienyl dehydratase, ... | | Authors: | Tan, K, Marshall, N, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-20 | | Release date: | 2009-11-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | The crystal structure of cyclohexadienyl dehydratase precursor from Pseudomonas aeruginosa PA01
To be Published
|
|
3Q94
 
 | | The crystal structure of fructose 1,6-bisphosphate aldolase from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, ACETATE ION, Fructose-bisphosphate aldolase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-07 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | The crystal structure of fructose 1,6-bisphosphate aldolase from Bacillus anthracis str. 'Ames Ancestor'.
To be Published
|
|
3L34
 
 | |
4DYU
 
 | | The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10 | | Descriptor: | DNA protection during starvation protein, SULFATE ION, ZINC ION | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10
To be Published
|
|
3KYZ
 
 | | The crystal structure of the sensor domain of two-component sensor PfeS from Pseudomonas aeruginosa PA01 | | Descriptor: | CHLORIDE ION, FORMIC ACID, Sensor protein pfeS | | Authors: | Tan, K, Marshall, N, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-07 | | Release date: | 2010-01-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | The crystal structure of the sensor domain of two-component sensor PfeS from Pseudomonas aeruginosa PA01
To be Published
|
|
3LDU
 
 | | The crystal structure of a possible methylase from Clostridium difficile 630. | | Descriptor: | FORMIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Tan, K, Wu, R, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-13 | | Release date: | 2010-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a possible methylase from Clostridium difficile 630.
To be Published
|
|
