2R5R
 
 | | The crystal structure of DUF198 from Nitrosomonas europaea ATCC 19718 | | Descriptor: | IMIDAZOLE, PHOSPHATE ION, UPF0343 protein NE1163 | | Authors: | Tan, K, Wu, R, Nocek, B, Bigelow, L, Patterson, S, Freeman, L, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The crystal structure of DUF198 from Nitrosomonas europaea ATCC 19718.
To be Published
|
|
2R8B
 
 | | The crystal structure of the protein Atu2452 of unknown function from Agrobacterium tumefaciens str. C58 | | Descriptor: | SULFATE ION, Uncharacterized protein Atu2452 | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-10 | | Release date: | 2007-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The crystal structure of the protein Atu2452 of unknown function from Agrobacterium tumefaciens str. C58.
TO BE PUBLISHED
|
|
2R5S
 
 | | The crystal structure of a domain of protein VP0806 (unknown function) from Vibrio parahaemolyticus RIMD 2210633 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Tan, K, Wu, R, Abdullah, J, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The crystal structure of a domain of protein VP0806 (unknown function) from Vibrio parahaemolyticus RIMD 2210633.
To be Published
|
|
4MA0
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with partially hydrolysed ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with partially hydrolysed ATP
To be Published
|
|
4MAM
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ADP analog, AMP-CP | | Descriptor: | GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Phosphoribosylaminoimidazole carboxylase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-16 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.474 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ADP analog, AMP-CP
To be Published
|
|
4M9U
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
4MA5
 
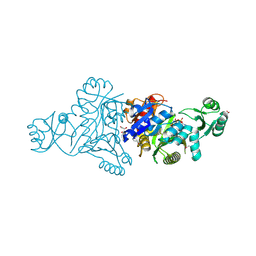 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ATP analog, AMP-PNP. | | Descriptor: | FORMIC ACID, GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.809 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ATP analog, AMP-PNP.
To be Published
|
|
4M9D
 
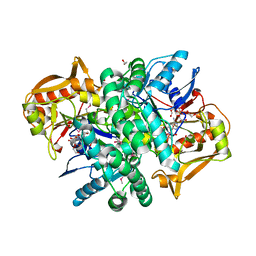 | | The Crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor in complex with AMP. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Adenylosuccinate synthetase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-14 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | The Crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor in complex with AMP.
To be Published
|
|
4M0C
 
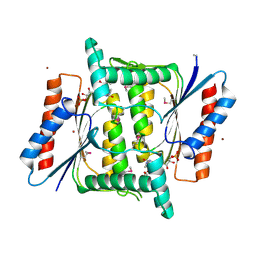 | | The crystal structure of a FMN-dependent NADH-azoreductase from Bacillus anthracis str. Ames Ancestor in complex with FMN. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, GLYCEROL, ... | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-01 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.073 Å) | | Cite: | The crystal structure of a FMN-dependent NADH-azoreductase from Bacillus anthracis str. Ames Ancestor in complex with FMN.
To be Published
|
|
4M0G
 
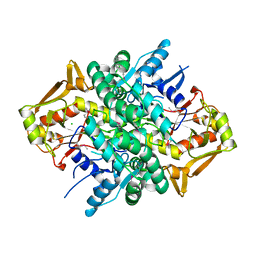 | | The crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor. | | Descriptor: | Adenylosuccinate synthetase, CHLORIDE ION | | Authors: | Tan, K, Zhou, M, Zhang, R, Kwon, K, Anderson, W.F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-01 | | Release date: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | The crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor.
To be Published
|
|
7KYU
 
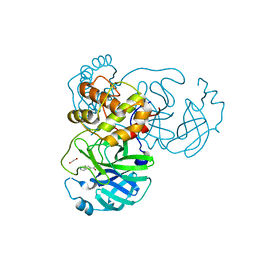 | | The crystal structure of SARS-CoV-2 Main Protease with the formation of Cys145-1H-indole-5-carboxylate | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1H-indole-5-carbonyl)oxy]-1H-benzotriazole, 3C-like proteinase | | Authors: | Tan, K, Maltseva, N.I, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The crystal structure of SARS-CoV-2 Main Protease with the formation of Cys145-1H-indole-5-carboxylate
To Be Published
|
|
4NEG
 
 | | The crystal structure of tryptophan synthase subunit beta from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | FORMIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Zhang, R, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-29 | | Release date: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | The crystal structure of tryptophan synthase subunit beta from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
4NPB
 
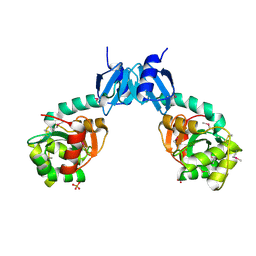 | | The crystal structure of thiol:disulfide interchange protein DsbC from Yersinia pestis CO92 | | Descriptor: | PHOSPHATE ION, Protein disulfide isomerase II, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-21 | | Release date: | 2013-12-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | The crystal structure of thiol:disulfide interchange protein DsbC from Yersinia pestis CO92
To be Published
|
|
4PZL
 
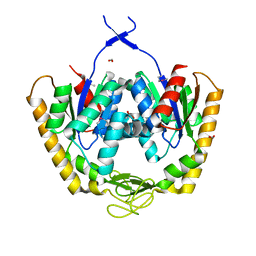 | | The crystal structure of adenylate kinase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | Adenylate kinase, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-03-31 | | Release date: | 2014-04-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of adenylate kinase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
4PZ0
 
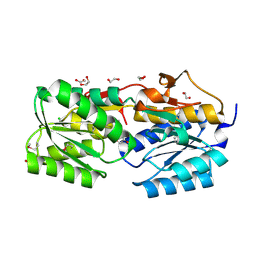 | | The crystal structure of a solute binding protein from Bacillus anthracis str. Ames in complex with quorum-sensing signal autoinducer-2 (AI-2) | | Descriptor: | (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-03-28 | | Release date: | 2014-04-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The crystal structure of a solute binding protein from Bacillus anthracis str. Ames in complex with quorum-sensing signal autoinducer-2 (AI-2).
To be Published
|
|
4R7Q
 
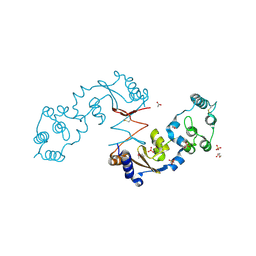 | | The structure of a sensor domain of a histidine kinase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-10 | | Last modified: | 2022-11-16 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 203, 2021
|
|
4RN7
 
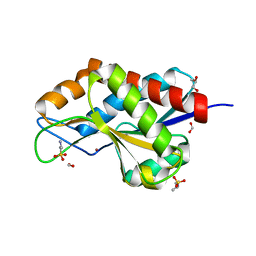 | | The crystal structure of N-acetylmuramoyl-L-alanine amidase from Clostridium difficile 630 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-23 | | Release date: | 2014-11-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | The crystal structure of N-acetylmuramoyl-L-alanine amidase from Clostridium difficile 630
To be Published
|
|
4S1N
 
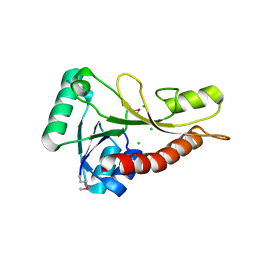 | | The crystal structure of phosphoribosylglycinamide formyltransferase from Streptococcus pneumoniae TIGR4 | | Descriptor: | CHLORIDE ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-01-14 | | Release date: | 2015-01-28 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of phosphoribosylglycinamide formyltransferase from Streptococcus pneumoniae TIGR4
To be Published
|
|
5ES2
 
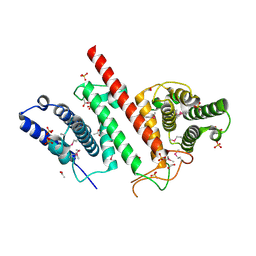 | | The crystal structure of a functionally uncharacterized protein LPG0634 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-11-16 | | Release date: | 2015-12-16 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of a functionally uncharacterized protein LPG0634 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To Be Published
|
|
5F4Z
 
 | | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus | | Descriptor: | (1~{R},2~{R})-2,3-dihydro-1~{H}-indene-1,2-diol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, BABNIGG, G, BINGMAN, C.A, YENNAMALLI, R, LOHMAN, J, Chang, C.Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-12-03 | | Release date: | 2016-02-17 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus
To Be Published
|
|
5EV7
 
 | | The crystal structure of a functionally unknown conserved protein mutant from Bacillus anthracis str. Ames | | Descriptor: | Conserved domain protein | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | The crystal structure of a functionally unknown conserved protein mutant from Bacillus anthracis str. Ames.
To Be Published
|
|
3UF6
 
 | | The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoD (3'-dephosphocoenzyme A) | | Descriptor: | DEPHOSPHO COENZYME A, Lmo1369 protein | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoD (3'-dephosphocoenzyme A)
To be Published
|
|
4DQD
 
 | | The crystal structure of a transporter in complex with 3-phenylpyruvic acid | | Descriptor: | 3-HYDROXYPYRUVIC ACID, 3-PHENYLPYRUVIC ACID, Extracellular ligand-binding receptor, ... | | Authors: | Tan, K, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-02-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4DQ0
 
 | |
4DZR
 
 | | The crystal structure of protein-(glutamine-N5) methyltransferase (release factor-specific) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Tan, K, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-03-01 | | Release date: | 2012-03-14 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | The crystal structure of protein-(glutamine-N5) methyltransferase (release factor-specific) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446
To be Published
|
|
