1N18
 
 | | Thermostable mutant of Human Superoxide Dismutase, C6A, C111S | | Descriptor: | COPPER (I) ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Cardoso, R.M.F, Thayer, M.M, DiDonato, M, Lo, T.P, Bruns, C.K, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2002-10-16 | | Release date: | 2002-11-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into Lou Gehrig's disease from the structure and instability of the A4V mutant of human Cu,Zn superoxide dismutase.
J.Mol.Biol., 324, 2002
|
|
1NOD
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER (DELTA 65) WITH TETRAHYDROBIOPTERIN AND SUBSTRATE L-ARGININE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1998-03-05 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of nitric oxide synthase oxygenase dimer with pterin and substrate.
Science, 279, 1998
|
|
1QB3
 
 | | CRYSTAL STRUCTURE OF THE CELL CYCLE REGULATORY PROTEIN CKS1 | | Descriptor: | CYCLIN-DEPENDENT KINASES REGULATORY SUBUNIT | | Authors: | Bourne, Y, Watson, M.H, Arvai, A.S, Bernstein, S.L, Reed, S.I, Tainer, J.A. | | Deposit date: | 1999-04-30 | | Release date: | 2000-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure and mutational analysis of the Saccharomyces cerevisiae cell cycle regulatory protein Cks1: implications for domain swapping, anion binding and protein interactions.
Structure Fold.Des., 8, 2000
|
|
1QYO
 
 | | Anaerobic precylization intermediate crystal structure for S65G Y66G GFP variant | | Descriptor: | green-fluorescent protein | | Authors: | Barondeau, D.P, Putnam, C.D, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2003-09-11 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism and energetics of green fluorescent protein chromophore synthesis revealed by trapped intermediate structures.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
1QXT
 
 | | Crystal structure of precyclized intermediate for the green fluorescent protein R96A variant (A) | | Descriptor: | green-fluorescent protein | | Authors: | Barondeau, D.P, Putnam, C.D, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2003-09-08 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism and energetics of green fluorescent protein chromophore synthesis revealed by trapped intermediate structures
Proc.Natl.Acad.Sci.USA, 100
|
|
1QY3
 
 | | Crystal structure of precyclized intermediate for the green fluorescent protein R96A variant (B) | | Descriptor: | green-fluorescent protein | | Authors: | Barondeau, D.P, Putnam, C.D, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2003-09-09 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism and energetics of green fluorescent protein chromophore synthesis revealed by trapped intermediate structures.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
1QNM
 
 | | HUMAN MANGANESE SUPEROXIDE DISMUTASE MUTANT Q143N | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Guan, Y, Tainer, J.A. | | Deposit date: | 1997-07-03 | | Release date: | 1998-01-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the active site of human manganese superoxide dismutase: the role of glutamine 143.
Biochemistry, 37, 1998
|
|
1QTW
 
 | | HIGH-RESOLUTION CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI DNA REPAIR ENZYME ENDONUCLEASE IV | | Descriptor: | ENDONUCLEASE IV, ZINC ION | | Authors: | Hosfield, D.J, Guan, Y, Haas, B.J, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 1999-06-29 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis.
Cell(Cambridge,Mass.), 98, 1999
|
|
1QYF
 
 | | Crystal structure of matured green fluorescent protein R96A variant | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, green-fluorescent protein | | Authors: | Barondeau, D.P, Putnam, C.D, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2003-09-10 | | Release date: | 2003-09-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism and energetics of green fluorescent protein chromophore synthesis revealed by trapped intermediate structures.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
1QYQ
 
 | | Crystal Structure of the cyclized S65G Y66G GFP variant | | Descriptor: | green-fluorescent protein | | Authors: | Barondeau, D.P, Putnam, C.D, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2003-09-11 | | Release date: | 2003-09-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism and energetics of green fluorescent protein chromophore synthesis revealed by trapped intermediate structures.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
1RWZ
 
 | | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) from A. fulgidus | | Descriptor: | DNA polymerase sliding clamp | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-17 | | Release date: | 2004-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
1RXM
 
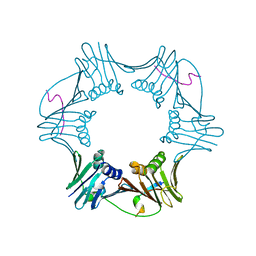 | | C-terminal region of FEN-1 bound to A. fulgidus PCNA | | Descriptor: | DNA polymerase sliding clamp, consensus FEN-1 peptide | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
1RXV
 
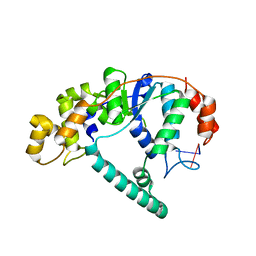 | | Crystal Structure of A. Fulgidus FEN-1 bound to DNA | | Descriptor: | 5'-d(*T*pA*pG*pC*pA*pT*pC*pG*pG), Flap structure-specific endonuclease | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
1RXW
 
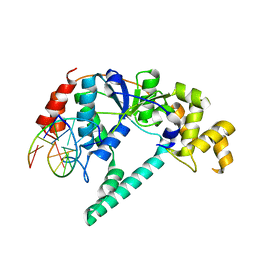 | | Crystal structure of A. fulgidus FEN-1 bound to DNA | | Descriptor: | 5'-d(*C*pG*pA*pT*pG*pC*pT*pA)-3', 5'-d(*T*pA*pG*pC*pA*pT*pC*pG*pG)-3', Flap structure-specific endonuclease | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
1RXZ
 
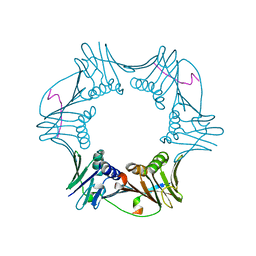 | | C-terminal region of A. fulgidus FEN-1 complexed with A. fulgidus PCNA | | Descriptor: | DNA polymerase sliding clamp, Flap structure-specific endonuclease | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
1SCE
 
 | |
1SOS
 
 | | ATOMIC STRUCTURES OF WILD-TYPE AND THERMOSTABLE MUTANT RECOMBINANT HUMAN CU, ZN SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE, ... | | Authors: | Parge, H.E, Hallewell, R.A, Tainer, J.A. | | Deposit date: | 1992-02-11 | | Release date: | 1993-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic structures of wild-type and thermostable mutant recombinant human Cu,Zn superoxide dismutase.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
1SPD
 
 | |
1T6U
 
 | | Nickel Superoxide Dismutase (NiSOD) Native 1.30 A Structure | | Descriptor: | NICKEL (II) ION, Superoxide dismutase [Ni] | | Authors: | Barondeau, D.P, Kassmann, C.J, Bruns, C.K, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2004-05-07 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Nickel superoxide dismutase structure and mechanism.
Biochemistry, 43, 2004
|
|
1T6Q
 
 | | Nickel Superoxide Dismutase (NiSOD) CN-treated Apo Structure | | Descriptor: | Superoxide dismutase [Ni] | | Authors: | Barondeau, D.P, Kassmann, C.J, Bruns, C.K, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2004-05-07 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Nickel superoxide dismutase structure and mechanism.
Biochemistry, 43, 2004
|
|
1TLL
 
 | | CRYSTAL STRUCTURE OF RAT NEURONAL NITRIC-OXIDE SYNTHASE REDUCTASE MODULE AT 2.3 A RESOLUTION. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Garcin, E.D, Bruns, C.M, Lloyd, S.J, Hosfield, D.J, Tiso, M, Gachhui, R, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2004-06-09 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase
J.Biol.Chem., 279, 2004
|
|
1T6I
 
 | | Nickel Superoxide Dismutase (NiSOD) Apo Structure | | Descriptor: | Superoxide dismutase [Ni] | | Authors: | Barondeau, D.P, Kassmann, C.J, Bruns, C.K, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2004-05-06 | | Release date: | 2004-07-13 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Nickel superoxide dismutase structure and mechanism.
Biochemistry, 43, 2004
|
|
1SZX
 
 | | Role Of Hydrogen Bonding In The Active Site Of Human Manganese Superoxide Dismutase | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Greenleaf, W.B, Perry, J.J, Hearn, A.S, Cabelli, D.E, Lepock, J.R, Stroupe, M.E, Tainer, J.A, Nick, H.S, Silverman, D.N. | | Deposit date: | 2004-04-06 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Role of hydrogen bonding in the active site of human manganese superoxide dismutase.
Biochemistry, 43, 2004
|
|
1UGI
 
 | | URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Descriptor: | IMIDAZOLE, SULFATE ION, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Putnam, C.D, Arvai, A.S, Mol, C.D, Tainer, J.A. | | Deposit date: | 1998-11-04 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
1UUG
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH WILD-TYPE UDG AND WILD-TYPE UGI | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Mol, C.D, Arvai, A.S, Putnam, C.D, Tainer, J.A. | | Deposit date: | 1998-10-31 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
