396D
 
 | |
319D
 
 | | CRYSTAL STRUCTURES OF D(CCGGG(BR)5CCCGG)-ORTHOGONAL FORM | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*GP*(CBR)P*CP*CP*GP*G)-3'), THERMINE | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
324D
 
 | | CRYSTAL STRUCTURES OF D(CCGGGCCM5CGG)-ORTHOGONAL FORM | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*GP*CP*CP*(5CM)P*GP*G)-3'), SPERMINE | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
377D
 
 | | 5'-R(*CP*GP*UP*AP*CP*DG)-3' | | Descriptor: | RNA-DNA (5'-R(*CP*GP*UP*AP*CP*DG)-3') | | Authors: | Biswas, R, Mitra, S.N, Sundaralingam, M. | | Deposit date: | 1998-01-26 | | Release date: | 1998-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | 1.76 A structure of a pyrimidine start alternating A-RNA hexamer r(CGUAC)dG.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
315D
 
 | |
325D
 
 | | CRYSTAL STRUCTURES OF D(CM5CGGGCCM5CGG)-HEXAGONAL FORM | | Descriptor: | DNA (5'-D(*CP*(5CM)P*GP*GP*GP*CP*CP*(5CM)P*GP*G)-3') | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
323D
 
 | | CRYSTAL STRUCTURES OF D(CCGGGCCM5CGG)-ORTHOGONAL FORM | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*GP*CP*CP*(5CM)P*GP*G)-3'), SPERMINE | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
375D
 
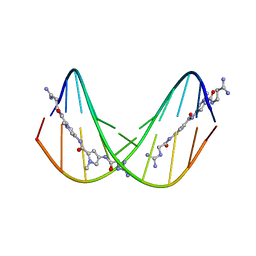 | | A NOVEL END-TO-END BINDING OF TWO NETROPSINS TO THE DNA DECAMER D(CCCCCIIIII)2 | | Descriptor: | DNA (5'-D(*CP*CP*CP*CP*CP*IP*IP*IP*IP*I)-3'), NETROPSIN | | Authors: | Chen, X, Rao, S.T, Sekar, K, Sundaralingam, M. | | Deposit date: | 1998-01-14 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Novel End-to-End Binding of Two Netropsins to the DNA Decamers d(CCCCCIIIII) 2, d(CCCBr5CCIIIII)2, d(CBr5CCCCIIIII)2
Nucleic Acids Res., 26, 1998
|
|
395D
 
 | |
3LYT
 
 | |
4LYT
 
 | |
2B96
 
 | | Third Calcium ion found in an inhibitor bound phospholipase A2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-METHOXYBENZOIC ACID, CALCIUM ION, ... | | Authors: | Sekar, K, Velmurugan, D, Yamane, T, Tsai, M.D. | | Deposit date: | 2005-10-11 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Third Calcium ion found in an inhibitor bound phospholipase A2
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BAX
 
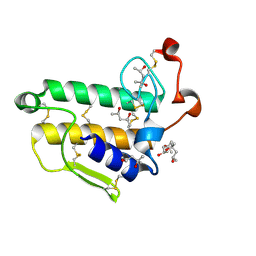 | | Atomic Resolution Structure of the Double Mutant (K53,56M) of Bovine Pancreatic Phospholipase A2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sekar, K, Yogavel, M, Velmurugan, D, Dauter, Z, Dauter, M, Tsai, M.D. | | Deposit date: | 2005-10-15 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution (0.97 A) structure of the triple mutant (K53,56,121M) of bovine pancreatic phospholipase A2.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1LKR
 
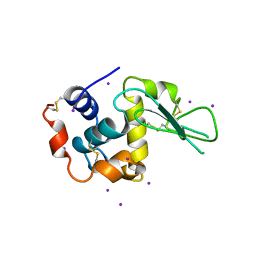 | | MONOCLINIC HEN EGG WHITE LYSOZYME IODIDE | | Descriptor: | IODIDE ION, LYSOZYME | | Authors: | Steinrauf, L.K. | | Deposit date: | 1998-01-20 | | Release date: | 1998-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of monoclinic lysozyme iodide at 1.6 A and of triclinic lysozyme nitrate at 1.1 A.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1ALC
 
 | |
1FLV
 
 | |
1LKS
 
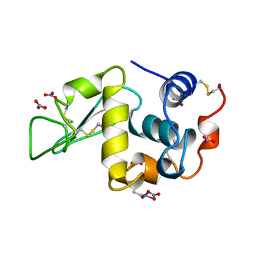 | | HEN EGG WHITE LYSOZYME NITRATE | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Steinrauf, L.K. | | Deposit date: | 1997-10-09 | | Release date: | 1998-04-29 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structures of monoclinic lysozyme iodide at 1.6 A and of triclinic lysozyme nitrate at 1.1 A.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1AZF
 
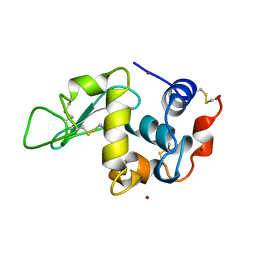 | | CHICKEN EGG WHITE LYSOZYME CRYSTAL GROWN IN BROMIDE SOLUTION | | Descriptor: | BROMIDE ION, LYSOZYME | | Authors: | Lim, K, Nadarajah, A, Forsythe, E.L, Pusey, M.L. | | Deposit date: | 1997-11-17 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Locations of bromide ions in tetragonal lysozyme crystals.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1EVV
 
 | |
1KOS
 
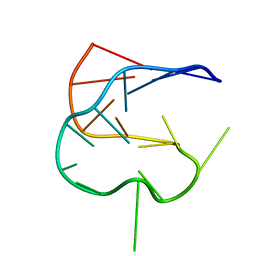 | | SOLUTION NMR STRUCTURE OF AN ANALOG OF THE YEAST TRNA PHE T STEM LOOP CONTAINING RIBOTHYMIDINE AT ITS NATURALLY OCCURRING POSITION | | Descriptor: | 5'-R(*CP*UP*GP*UP*GP*(5MU)P*UP*CP*GP*AP*UP*(CH)P*CP*AP*CP*AP*G)- 3' | | Authors: | Koshlap, K.M, Guenther, R, Sochacka, E, Malkiewicz, A, Agris, P.F. | | Deposit date: | 1999-05-03 | | Release date: | 1999-10-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A distinctive RNA fold: the solution structure of an analogue of the yeast tRNAPhe T Psi C domain.
Biochemistry, 38, 1999
|
|
