6L85
 
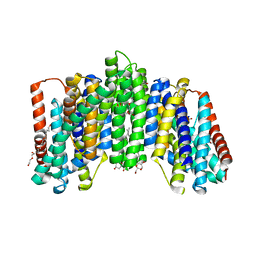 | | The sodium-dependent phosphate transporter | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-BETA-D-MALTOSIDE, PHOSPHATE ION, ... | | Authors: | Tsai, J.-Y, Sun, Y.-J. | | Deposit date: | 2019-11-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structure of the sodium-dependent phosphate transporter reveals insights into human solute carrier SLC20.
Sci Adv, 6, 2020
|
|
2BQX
 
 | |
2CCZ
 
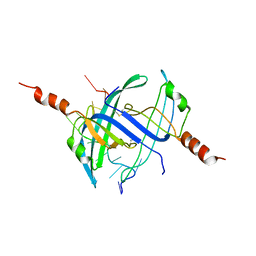 | | Crystal structure of E. coli primosomol protein PriB bound to ssDNA | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP *TP*TP*TP*TP*T)-3', PRIMOSOMAL REPLICATION PROTEIN N | | Authors: | Huang, C.-Y, Hsu, C.-H, Wu, H.-N, Sun, Y.-J, Hsiao, C.-D. | | Deposit date: | 2006-01-19 | | Release date: | 2006-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complexed Crystal Structure of Replication Restart Primsome Protein Prib Reveals a Novel Single-Stranded DNA-Binding Mode.
Nucleic Acids Res., 34, 2006
|
|
2BQY
 
 | |
2C6Y
 
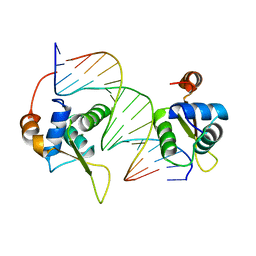 | | Crystal structure of interleukin enhancer-binding factor 1 bound to DNA | | Descriptor: | FORKHEAD BOX PROTEIN K2, INTERLEUKIN 2 PROMOTOR, MAGNESIUM ION | | Authors: | Tsai, K.-L, Huang, C.-Y, Chang, C.-H, Sun, Y.-J, Chuang, W.-J, Hsiao, C.-D. | | Deposit date: | 2005-11-15 | | Release date: | 2006-04-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Human Foxk1A-DNA Complex and its Implications on the Diverse Binding Specificity of Winged Helix/Forkhead Proteins.
J.Biol.Chem., 281, 2006
|
|
1WDN
 
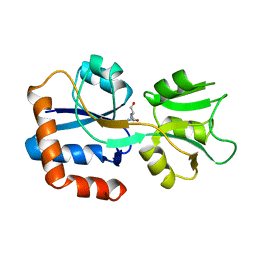 | | GLUTAMINE-BINDING PROTEIN | | Descriptor: | GLUTAMINE, GLUTAMINE BINDING PROTEIN | | Authors: | Sun, Y.-J, Rose, J, Wang, B.-C, Hsiao, C.-D. | | Deposit date: | 1997-05-17 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The structure of glutamine-binding protein complexed with glutamine at 1.94 A resolution: comparisons with other amino acid binding proteins.
J.Mol.Biol., 278, 1998
|
|
1GSU
 
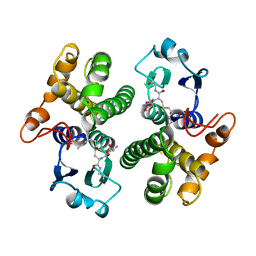 | | AN AVIAN CLASS-MU GLUTATHIONE S-TRANSFERASE, CGSTM1-1 AT 1.94 ANGSTROM RESOLUTION | | Descriptor: | CLASS-MU GLUTATHIONE S-TRANSFERASE, S-HEXYLGLUTATHIONE | | Authors: | Sun, Y.-J, Kuan, C, Tam, M.F, Hsiao, C.-D. | | Deposit date: | 1997-09-02 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The three-dimensional structure of an avian class-mu glutathione S-transferase, cGSTM1-1 at 1.94 A resolution.
J.Mol.Biol., 278, 1998
|
|
1KXI
 
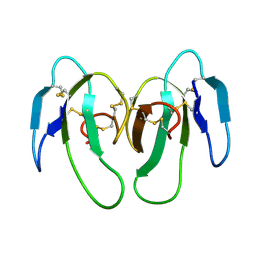 | | STRUCTURE OF CYTOTOXIN HOMOLOG PRECURSOR | | Descriptor: | CARDIOTOXIN V | | Authors: | Sun, Y.-J, Wu, W.-G, Chiang, C.-M, Hsin, A.-Y, Hsiao, C.-D. | | Deposit date: | 1996-08-29 | | Release date: | 1997-04-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of cardiotoxin V from Taiwan cobra venom: pH-dependent conformational change and a novel membrane-binding motif identified in the three-finger loops of P-type cardiotoxin.
Biochemistry, 36, 1997
|
|
2PGI
 
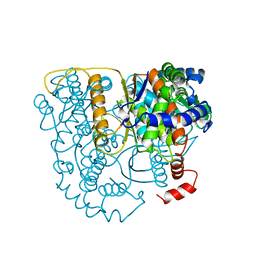 | | THE CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE-AN ENZYME WITH AUTOCRINE MOTILITY FACTOR ACTIVITY IN TUMOR CELLS | | Descriptor: | PHOSPHOGLUCOSE ISOMERASE | | Authors: | Sun, Y.-J, Chou, C.-C, Chen, W.-S, Meng, M, Hsiao, C.-D. | | Deposit date: | 1998-10-27 | | Release date: | 1999-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of a multifunctional protein: phosphoglucose isomerase/autocrine motility factor/neuroleukin.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1GGG
 
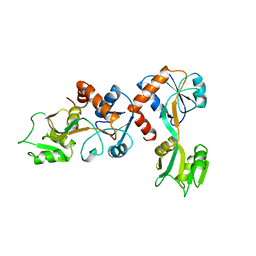 | |
4CXO
 
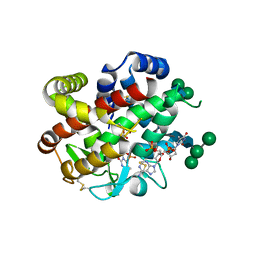 | | bifunctional endonuclease in complex with ssDNA | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, ENDONUCLEASE 2, SULFATE ION, ... | | Authors: | Yu, T.-F, Maestre-Reyna, M, Ko, C.-Y, Ko, T.-P, Sun, Y.-J, Lin, T.-Y, Shaw, J.-F, Wang, A.H.-J. | | Deposit date: | 2014-04-08 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Insights of the Ssdna Binding Site in the Multifunctional Endonuclease Atbfn2 from Arabidopsis Thaliana.
Plos One, 9, 2014
|
|
4CXV
 
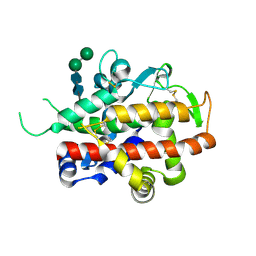 | | Structure of bifunctional endonuclease (AtBFN2) in complex with phosphate. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDONUCLEASE 2, ... | | Authors: | Yu, T.-F, Maestre-Reyna, M, Ko, C.-Y, Ko, T.-P, Sun, Y.-J, Lin, T.-Y, Shaw, J.-F, Wang, A.H.-J. | | Deposit date: | 2014-04-09 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights of the Ssdna Binding Site in the Multifunctional Endonuclease Atbfn2 from Arabidopsis Thaliana.
Plos One, 9, 2014
|
|
4CXP
 
 | | Structure of bifunctional endonuclease (AtBFN2) from Arabidopsis thaliana in complex with sulfate | | Descriptor: | ENDONUCLEASE 2, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Yu, T.-F, Maestre-Reyna, M, Ko, C.-Y, Ko, T.-P, Sun, Y.-J, Lin, T.-Y, Shaw, J.-F, Wang, A.H.-J. | | Deposit date: | 2014-04-08 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structural Insights of the Ssdna Binding Site in the Multifunctional Endonuclease Atbfn2 from Arabidopsis Thaliana.
Plos One, 9, 2014
|
|
4CWM
 
 | | High-glycosylation crystal structure of the bifunctional endonuclease (AtBFN2) from Arabidopsis thaliana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDONUCLEASE 2, ZINC ION, ... | | Authors: | Yu, T.-F, Maestre-Reyna, M, Ko, C.-Y, Ko, T.-P, Sun, Y.-J, Lin, T.-Y, Shaw, J.-F, Wang, A.H.-J. | | Deposit date: | 2014-04-03 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Insights of the Ssdna Binding Site in the Multifunctional Endonuclease Atbfn2 from Arabidopsis Thaliana.
Plos One, 9, 2014
|
|
2VN2
 
 | | Crystal structure of the N-terminal domain of DnaD protein from Geobacillus kaustophilus HTA426 | | Descriptor: | CHROMOSOME REPLICATION INITIATION PROTEIN, MAGNESIUM ION | | Authors: | Huang, C.-Y, Chang, Y.-W, Chen, W.-T, Sun, Y.-J, Hsiao, C.-D. | | Deposit date: | 2008-01-30 | | Release date: | 2008-08-12 | | Last modified: | 2012-08-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the N-Terminal Domain of Geobacillus Kaustophilus Hta426 Dnad Protein.
Biochem.Biophys.Res.Commun., 375, 2008
|
|
6AFZ
 
 | | Proton pyrophosphatase-E225H mutant | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tsai, J.-Y, Li, K.-M, Sun, Y.-J. | | Deposit date: | 2018-08-08 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Roles of the Hydrophobic Gate and Exit Channel in Vigna radiata Pyrophosphatase Ion Translocation.
J. Mol. Biol., 431, 2019
|
|
6AFW
 
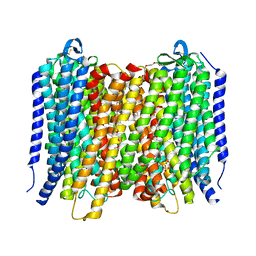 | | Proton pyrophosphatase-T228D mutant | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tsai, J.-Y, Li, K.-M, Sun, Y.-J. | | Deposit date: | 2018-08-08 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | Roles of the Hydrophobic Gate and Exit Channel in Vigna radiata Pyrophosphatase Ion Translocation.
J. Mol. Biol., 431, 2019
|
|
6AFV
 
 | | Proton pyrophosphatase-L555K mutant | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tsai, J.-Y, Li, K.-M, Sun, Y.-J. | | Deposit date: | 2018-08-08 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Roles of the Hydrophobic Gate and Exit Channel in Vigna radiata Pyrophosphatase Ion Translocation.
J. Mol. Biol., 431, 2019
|
|
6AFT
 
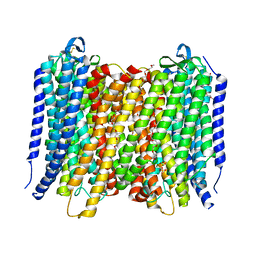 | | Proton pyrophosphatase - E301Q | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tsai, J.-Y, Tang, K.-Z, Sun, Y.-J. | | Deposit date: | 2018-08-08 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Roles of the Hydrophobic Gate and Exit Channel in Vigna radiata Pyrophosphatase Ion Translocation.
J. Mol. Biol., 431, 2019
|
|
6AFU
 
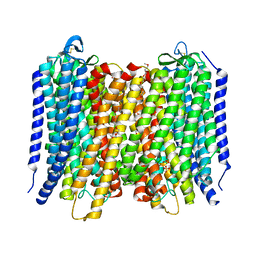 | | Proton pyrophosphatase-L555M mutant | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tsai, J.-Y, Li, K.-M, Sun, Y.-J. | | Deposit date: | 2018-08-08 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Roles of the Hydrophobic Gate and Exit Channel in Vigna radiata Pyrophosphatase Ion Translocation.
J. Mol. Biol., 431, 2019
|
|
6AFY
 
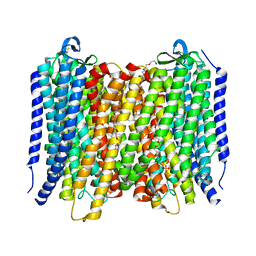 | | Proton pyrophosphatase-E225S mutant | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tsai, J.-Y, Li, K.-M, Sun, Y.-J. | | Deposit date: | 2018-08-08 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Roles of the Hydrophobic Gate and Exit Channel in Vigna radiata Pyrophosphatase Ion Translocation.
J. Mol. Biol., 431, 2019
|
|
6AFS
 
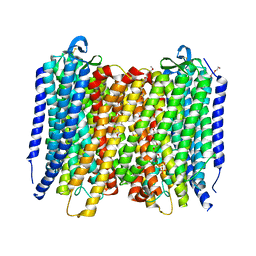 | | Proton pyrophosphatase - two phosphates-bound | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tsai, J.-Y, Li, K.-M, Sun, Y.-J. | | Deposit date: | 2018-08-08 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Roles of the Hydrophobic Gate and Exit Channel in Vigna radiata Pyrophosphatase Ion Translocation.
J. Mol. Biol., 431, 2019
|
|
6AFX
 
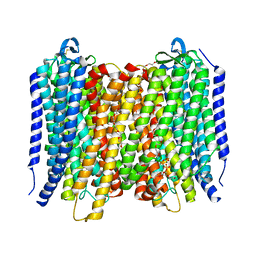 | | Proton pyrophosphatase - E225A | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tsai, J.-Y, Tang, K.-Z, Sun, Y.-J. | | Deposit date: | 2018-08-08 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Roles of the Hydrophobic Gate and Exit Channel in Vigna radiata Pyrophosphatase Ion Translocation.
J. Mol. Biol., 431, 2019
|
|
2J3E
 
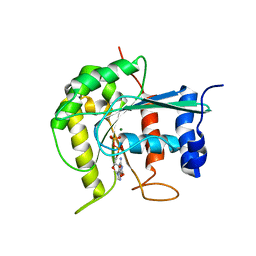 | | Dimerization is important for the GTPase activity of chloroplast translocon components atToc33 and psToc159 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, T7I23.11 PROTEIN | | Authors: | Yeh, Y.-H, Kesavulu, M.M, Wu, S.-Z, Li, H.-M, Sun, Y.-J, Konozy, E.H, Hsiao, C.-D. | | Deposit date: | 2006-08-21 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dimerization is Important for the Gtpase Activity of Chloroplast Translocon Components Attoc33 and Pstoc159.
J.Biol.Chem., 282, 2007
|
|
