2F0C
 
 | |
4RGA
 
 | | Phage 1358 receptor binding protein in complex with the trisaccharide GlcNAc-Galf-GlcOMe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-galactofuranose-(1-6)-methyl alpha-D-glucopyranoside, Phage 1358 receptor binding protein (ORF20) | | Authors: | Spinelli, S, Mccabe, O, Farenc, C, Tremblay, D, Blangy, S, Oscarson, S, Moineau, S, Cambillau, C. | | Deposit date: | 2014-09-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The targeted recognition of Lactococcus lactis phages to their polysaccharide receptors.
Mol.Microbiol., 96, 2015
|
|
3EJC
 
 | | Full length Receptor Binding Protein from Lactococcal phage TP901-1 | | Descriptor: | Baseplate protein (BPP) | | Authors: | Spinelli, S, Lichiere, J, Blangy, S, Sciara, G, Cambillau, C, Campanacci, V. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and molecular assignment of lactococcal phage TP901-1 baseplate.
J.Biol.Chem., 285, 2010
|
|
2HLV
 
 | | Bovine Odorant Binding Protein deswapped triple mutant | | Descriptor: | 3,6-BIS(METHYLENE)DECANOIC ACID, GLYCEROL, Odorant-binding protein | | Authors: | Spinelli, S, Tegoni, M, Cambillau, C. | | Deposit date: | 2006-07-10 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Deswapping bovine odorant binding protein.
Biochim.Biophys.Acta, 1784, 2008
|
|
3PM2
 
 | | Crystal structure of a novel type of odorant binding protein from Anopheles gambiae belonging to the c+ class | | Descriptor: | Odorant binding protein (AGAP007287-PA) | | Authors: | Spinelli, S, Lagarde, A, Qiao, H, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2010-11-16 | | Release date: | 2011-05-25 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a novel type of odorant-binding protein from Anopheles gambiae, belonging to the C-plus class.
Biochem.J., 437, 2011
|
|
4V96
 
 | | The structure of a 1.8 MDa viral genome injection device suggests alternative infection mechanisms | | Descriptor: | BPP, ORF46, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V, van Sinderen, D, Cambillau, C. | | Deposit date: | 2012-02-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5NGH
 
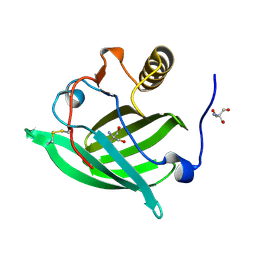 | |
3HG0
 
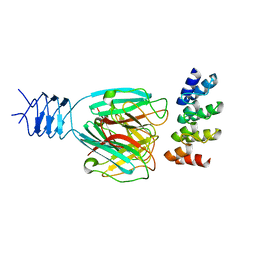 | | Crystal structure of a DARPin in complex with ORF49 from Lactococcal phage TP901-1 | | Descriptor: | Baseplate protein, Designed Ankyrin Repeat Protein (DARPin) 20 | | Authors: | Veesler, D, Dreier, B, Blangy, S, Lichiere, J, Tremblay, D, Moineau, S, Spinelli, S, Tegoni, M, Pluckthun, A, Campanacci, V, Cambillau, C. | | Deposit date: | 2009-05-13 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and function of a DARPin neutralizing inhibitor of lactococcal phage TP901-1: comparison of DARPin and camelid VHH binding mode.
J.Biol.Chem., 284, 2009
|
|
2H36
 
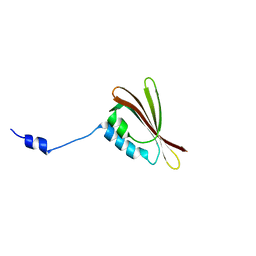 | |
3FE6
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera with a serendipitous ligand at pH 5.5 | | Descriptor: | (20S)-20-methyldotetracontane, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-11-27 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Queen bee pheromone binding protein pH induced domain-swapping favors pheromone release
To be Published
|
|
3FE9
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera with a serendipitous ligand soaked at pH 7.0 | | Descriptor: | (20S)-20-methyldotetracontane, CHLORIDE ION, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Queen bee pheromone binding protein pH induced domain-swapping favors pheromone release
To be Published
|
|
3FE8
 
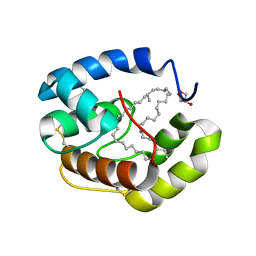 | | Crystal structure of a pheromone binding protein from Apis mellifera with a serendipitous ligand soaked at pH 4.0 | | Descriptor: | (20S)-20-methyldotetracontane, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Queen bee pheromone binding protein pH induced domain-swapping favors pheromone release
To be Published
|
|
1XYZ
 
 | |
5E7F
 
 | | Complex between lactococcal phage Tuc2009 RBP head domain and a nanobody (L06) | | Descriptor: | Major structural protein 1, nanobody L06 | | Authors: | Legrand, P, Collins, B, Blangy, S, Murphy, J, Spinelli, S, Gutierrez, C, Richet, N, Kellenberger, C, Desmyter, A, Mahony, J, van Sinderen, D, Cambillau, C. | | Deposit date: | 2015-10-12 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Mbio, 7, 2016
|
|
5E7T
 
 | | Structure of the tripod (BppUct-A-L) from the baseplate of bacteriophage Tuc2009 | | Descriptor: | CALCIUM ION, Major structural protein 1, Minor structural protein 4, ... | | Authors: | Legrand, P, Collins, B, Blangy, S, Murphy, J, Spinelli, S, Gutierrez, C, Richet, N, Kellenberger, C, Desmyter, A, Mahony, J, van Sinderen, D, Cambillau, C. | | Deposit date: | 2015-10-13 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Mbio, 7, 2016
|
|
5E7B
 
 | | Structure of a nanobody (vHH) from camel against phage Tuc2009 RBP (BppL, ORF53) | | Descriptor: | nanobody nano-L06 | | Authors: | Legrand, P, Collins, B, Blangy, S, Murphy, J, Spinelli, S, Gutierrez, C, Richet, N, Kellenberger, C, Desmyter, A, Mahony, J, van Sinderen, D, Cambillau, C. | | Deposit date: | 2015-10-12 | | Release date: | 2015-12-30 | | Last modified: | 2016-05-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Mbio, 7, 2016
|
|
4L97
 
 | | Structure of the RBP of lactococcal phage 1358 in complex with glucose-1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, Receptor Binding Protein | | Authors: | Farenc, C, Spinelli, S, Bebeacua, C, Tremblay, D, Orlov, I, Blangy, S, Klaholz, B.P, Moineau, S, Cambillau, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | A Virulent Siphophage CyoEM Structure and Host Recognition and Infection Mechanism
To be Published
|
|
4L92
 
 | | Structure of the RBP from lactococcal phage 1358 in complex with 2 GlcNAc molecules | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor Binding Protein, ... | | Authors: | Farenc, C, Spinelli, S, Bebeacua, C, Tremblay, D, Orlov, I, Blangy, S, Klaholz, B.P, Moineau, S, Cambillau, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Virulent Siphophage CyoEM Structure and Host Recognition and Infection Mechanism
To be Published
|
|
4L9B
 
 | | Structure of native RBP from lactococcal phage 1358 (CsI derivative) | | Descriptor: | CESIUM ION, Receptor Binding Protein | | Authors: | Farenc, C, Spinelli, S, Bebeacua, C, Tremblay, D, Orlov, I, Blangy, S, Klaholz, B.P, Moineau, S, Cambillau, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Virulent Siphophage CyoEM Structure and Host Recognition and Infection Mechanism
To be Published
|
|
4L99
 
 | | Structure of the RBP from lactococcal phage 1358 in complex with glycerol | | Descriptor: | GLYCEROL, Receptor Binding Protein, ZINC ION | | Authors: | Farenc, C, Spinelli, S, Bebeacua, C, Tremblay, D, Orlov, I, Blangy, S, Klaholz, B.P, Moineau, S, Cambillau, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Virulent Siphophage CyoEM Structure and Host Recognition and Infection Mechanism
To be Published
|
|
3DJW
 
 | | The thermo- and acido-stable ORF-99 from the archaeal virus AFV1 | | Descriptor: | ORF99 | | Authors: | Goulet, A, Spinelli, S, Prangishvili, D, van Tilbeurgh, H, Cambillau, C, Campanacci, V. | | Deposit date: | 2008-06-24 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The thermo- and acido-stable ORF-99 from the archaeal virus AFV1
Protein Sci., 18, 2009
|
|
5EFV
 
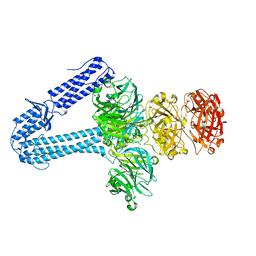 | | The host-recognition device of Staphylococcus aureus phage Phi11 | | Descriptor: | FE (III) ION, Phi ETA orf 56-like protein | | Authors: | Koc, C, Kuhner, P, Xia, G, Spinelli, S, Roussel, A, Cambillau, C, Stehle, T. | | Deposit date: | 2015-10-26 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the host-recognition device of Staphylococcus aureus phage 11.
Sci Rep, 6, 2016
|
|
4YO3
 
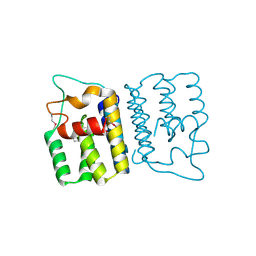 | | Enteroaggregative Escherichia Coli TssA N-terminal fragment | | Descriptor: | TssA | | Authors: | Durand, E, Zoued, A, Spinelli, S, Douzi, B, Brunet, Y.R, Bebeacua, C, Legrand, P, Journet, L, Mignot, T, Cambillau, C, Cascales, E. | | Deposit date: | 2015-03-11 | | Release date: | 2016-02-17 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Priming and polymerization of a bacterial contractile tail structure.
Nature, 531, 2016
|
|
4Y7M
 
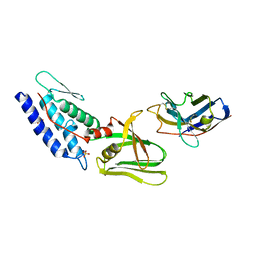 | | T6SS protein TssM C-terminal domain (835-1129) from EAEC | | Descriptor: | Hi113 protein, SULFATE ION, Type VI secretion protein IcmF | | Authors: | Nguyen, V.S, Spinelli, S, Durand, E, Roussel, A, Cambillau, C. | | Deposit date: | 2015-02-15 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biogenesis and structure of a type VI secretion membrane core complex.
Nature, 523, 2015
|
|
4Y7L
 
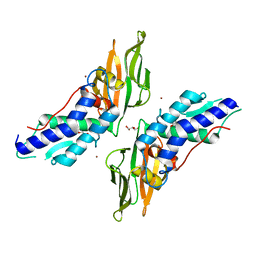 | | T6SS protein TssM C-terminal domain (869-1107) from EAEC | | Descriptor: | GLYCEROL, Type VI secretion protein IcmF, ZINC ION | | Authors: | Nguyen, V.S, Spinelli, S, Durand, E, Roussel, A, Cambillau, C. | | Deposit date: | 2015-02-15 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Biogenesis and structure of a type VI secretion membrane core complex.
Nature, 523, 2015
|
|
