1GUH
 
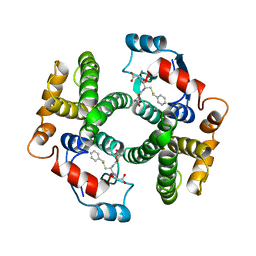 | | Structure determination and refinement of human alpha class glutathione transferase A1-1, and a comparison with the MU and PI class enzymes | | 分子名称: | GLUTATHIONE S-TRANSFERASE A1-1, S-BENZYL-GLUTATHIONE | | 著者 | Sinning, I, Kleywegt, G.J, Jones, T.A. | | 登録日 | 1993-02-24 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure determination and refinement of human alpha class glutathione transferase A1-1, and a comparison with the Mu and Pi class enzymes.
J.Mol.Biol., 232, 1993
|
|
1SUR
 
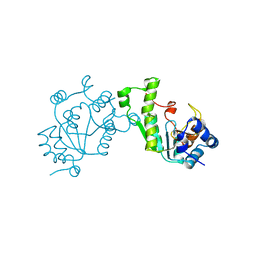 | | PHOSPHO-ADENYLYL-SULFATE REDUCTASE | | 分子名称: | PAPS REDUCTASE | | 著者 | Sinning, I, Savage, H. | | 登録日 | 1998-04-01 | | 公開日 | 1999-05-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of phosphoadenylyl sulphate (PAPS) reductase: a new family of adenine nucleotide alpha hydrolases.
Structure, 5, 1997
|
|
4V7F
 
 | | Arx1 pre-60S particle. | | 分子名称: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | 著者 | Leidig, C, Thoms, M, Holdermann, I, Bradatsch, B, Berninghausen, O, Bange, G, Sinning, I, Hurt, E, Beckmann, R. | | 登録日 | 2013-12-10 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (8.7 Å) | | 主引用文献 | 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle.
Nat Commun, 5, 2014
|
|
4WJS
 
 | |
4WJV
 
 | | Crystal structure of Rsa4 in complex with the Nsa2 binding peptide | | 分子名称: | Maltose-binding periplasmic protein, Ribosome assembly protein 4, Ribosome biogenesis protein NSA2, ... | | 著者 | Holdermann, I, Paternoga, H, Bassler, J, Hurt, E, Sinning, I. | | 登録日 | 2014-10-01 | | 公開日 | 2014-11-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
4WFM
 
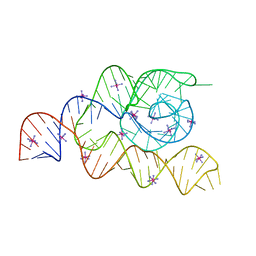 | | Structure of the complete bacterial SRP Alu domain | | 分子名称: | Bacillus subtilis small cytoplasmic RNA (scRNA),RNA, COBALT HEXAMMINE(III), MAGNESIUM ION | | 著者 | Kempf, G, Wild, K, Sinning, I. | | 登録日 | 2014-09-15 | | 公開日 | 2014-10-15 | | 最終更新日 | 2017-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of the complete bacterial SRP Alu domain.
Nucleic Acids Res., 42, 2014
|
|
8CR1
 
 | | Homo sapiens Get1/Get2 heterotetramer in complex with a Get3 dimer | | 分子名称: | ATPase ASNA1, Guided entry of tail-anchored proteins factor CAMLG,Guided entry of tail-anchored proteins factor 1,GET2-GET1, ZINC ION | | 著者 | McDowell, M.A, Heimes, M, Wild, K, Sinning, I. | | 登録日 | 2023-03-07 | | 公開日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
7OJU
 
 | | Chaetomium thermophilum Naa50 GNAT-domain in complex with bisubstrate analogue CoA-Ac-MVNAL | | 分子名称: | CARBOXYMETHYL COENZYME *A, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | 著者 | Weidenhausen, J, Kopp, J, Sinning, I. | | 登録日 | 2021-05-17 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Extended N-Terminal Acetyltransferase Naa50 in Filamentous Fungi Adds to Naa50 Diversity.
Int J Mol Sci, 23, 2022
|
|
8CQZ
 
 | | Homo sapiens Get3 in complex with the Get1 cytoplasmic domain | | 分子名称: | ATPase ASNA1, Guided entry of tail-anchored proteins factor 1 | | 著者 | McDowell, M.A, Heimes, M, Wild, K, Saar, D, Sinning, I. | | 登録日 | 2023-03-07 | | 公開日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
8CR2
 
 | | Homo sapiens Get1/Get2 heterotetramer (a3' deletion variant) in complex with a Get3 dimer | | 分子名称: | ATPase ASNA1, Guided entry of tail-anchored proteins factor CAMLG,Guided entry of tail-anchored proteins factor 1, ZINC ION | | 著者 | McDowell, M.A, Heimes, M, Wild, K, Sinning, I. | | 登録日 | 2023-03-07 | | 公開日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
8ODV
 
 | | Chaetomium thermophilum Get1/Get2 heterotetramer in complex with a Get3 dimer (nanodisc) | | 分子名称: | ATPase GET3, Protein GET2,Protein GET1, ZINC ION | | 著者 | McDowell, M.A, Wild, K, Sinning, I. | | 登録日 | 2023-03-09 | | 公開日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
8ODU
 
 | | Chaetomium thermophilum Get1/Get2 heterotetramer in complex with a Get3 dimer (amphipol) | | 分子名称: | ATPase GET3, Protein GET2,Protein GET1, ZINC ION | | 著者 | McDowell, M.A, Wild, K, Sinning, I. | | 登録日 | 2023-03-09 | | 公開日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
7Q72
 
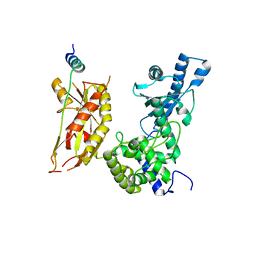 | | Structure of Pla1 in complex with Red1 | | 分子名称: | NURS complex subunit red1, Poly(A) polymerase pla1 | | 著者 | Soni, K, Wild, K, Sinning, I. | | 登録日 | 2021-11-09 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-04-05 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex.
Nat Commun, 14, 2023
|
|
5NZS
 
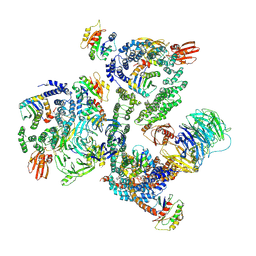 | | The structure of the COPI coat leaf in complex with the ArfGAP2 uncoating factor | | 分子名称: | ADP-ribosylation factor 1, ADP-ribosylation factor GTPase-activating protein 2, Coatomer subunit alpha, ... | | 著者 | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | 登録日 | 2017-05-15 | | 公開日 | 2017-06-28 | | 最終更新日 | 2018-10-24 | | 実験手法 | ELECTRON MICROSCOPY (10.1 Å) | | 主引用文献 | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5NZR
 
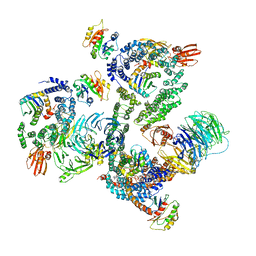 | | The structure of the COPI coat leaf | | 分子名称: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | 著者 | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | 登録日 | 2017-05-15 | | 公開日 | 2017-06-28 | | 最終更新日 | 2018-10-24 | | 実験手法 | ELECTRON MICROSCOPY (9.2 Å) | | 主引用文献 | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5NZU
 
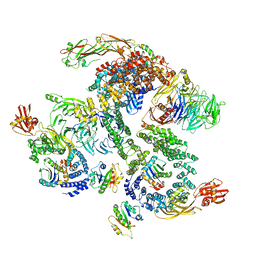 | | The structure of the COPI coat linkage II | | 分子名称: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | 著者 | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | 登録日 | 2017-05-15 | | 公開日 | 2017-06-28 | | 最終更新日 | 2018-10-24 | | 実験手法 | ELECTRON MICROSCOPY (15 Å) | | 主引用文献 | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5NZT
 
 | | The structure of the COPI coat linkage I | | 分子名称: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | 著者 | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | 登録日 | 2017-05-15 | | 公開日 | 2017-06-28 | | 最終更新日 | 2018-10-24 | | 実験手法 | ELECTRON MICROSCOPY (17 Å) | | 主引用文献 | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5NZV
 
 | | The structure of the COPI coat linkage IV | | 分子名称: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | 著者 | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | 登録日 | 2017-05-15 | | 公開日 | 2017-06-28 | | 最終更新日 | 2018-10-24 | | 実験手法 | ELECTRON MICROSCOPY (17.299999 Å) | | 主引用文献 | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
7OLC
 
 | | Thermophilic eukaryotic 80S ribosome at idle POST state | | 分子名称: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | 著者 | Kisonaite, M, Wild, K, Sinning, I. | | 登録日 | 2021-05-19 | | 公開日 | 2022-01-26 | | 最終更新日 | 2022-04-06 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | High-resolution structures of a thermophilic eukaryotic 80S ribosome reveal atomistic details of translocation.
Nat Commun, 13, 2022
|
|
7OLD
 
 | | Thermophilic eukaryotic 80S ribosome at pe/E (TI)-POST state | | 分子名称: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | 著者 | Kisonaite, M, Wild, K, Sinning, I. | | 登録日 | 2021-05-19 | | 公開日 | 2022-01-26 | | 最終更新日 | 2022-02-09 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | High-resolution structures of a thermophilic eukaryotic 80S ribosome reveal atomistic details of translocation.
Nat Commun, 13, 2022
|
|
1FTS
 
 | |
4WJU
 
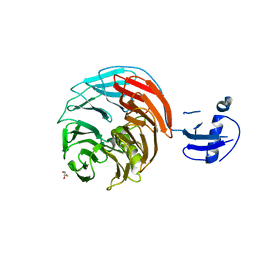 | | Crystal structure of Rsa4 from Saccharomyces cerevisiae | | 分子名称: | GLYCEROL, Ribosome assembly protein 4 | | 著者 | Holdermann, I, Bassler, J, Hurt, E, Sinning, I. | | 登録日 | 2014-10-01 | | 公開日 | 2014-11-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
4ADU
 
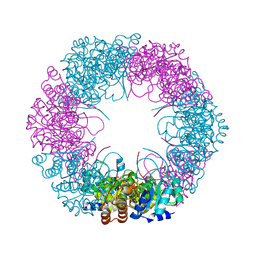 | | Crystal structure of plasmodial PLP synthase with bound R5P intermediate | | 分子名称: | 1,2-ETHANEDIOL, PYRIDOXINE BIOSYNTHETIC ENZYME PDX1 HOMOLOGUE, PUTATIVE, ... | | 著者 | Guedez, G, Sinning, I, Tews, I. | | 登録日 | 2012-01-03 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Assembly of the Eukaryotic Plp-Synthase Complex from Plasmodium and Activation of the Pdx1 Enzyme.
Structure, 20, 2012
|
|
4ADS
 
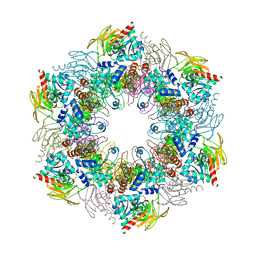 | | Crystal structure of plasmodial PLP synthase complex | | 分子名称: | PDX2 PROTEIN, PHOSPHATE ION, PYRIDOXINE BIOSYNTHETIC ENZYME PDX1 HOMOLOGUE, ... | | 著者 | Guedez, G, Sinning, I, Tews, I. | | 登録日 | 2012-01-03 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.61 Å) | | 主引用文献 | Assembly of the Eukaryotic Plp-Synthase Complex from Plasmodium and Activation of the Pdx1 Enzyme.
Structure, 20, 2012
|
|
4ADT
 
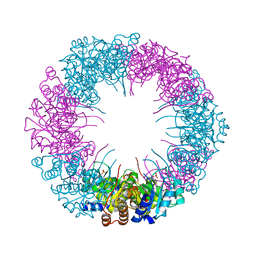 | |
