4EGJ
 
 | |
4GNV
 
 | |
3OMF
 
 | |
5UZH
 
 | |
3NDO
 
 | |
3NG3
 
 | |
5W15
 
 | |
5VPQ
 
 | |
5SCO
 
 | |
5SD3
 
 | |
5SCV
 
 | |
5SCR
 
 | |
6PZJ
 
 | |
6U3L
 
 | |
6XK2
 
 | |
6XR5
 
 | |
6X79
 
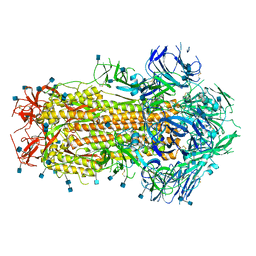 | | Prefusion SARS-CoV-2 S ectodomain trimer covalently stabilized in the closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | McCallum, M, Walls, A.C, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-05-29 | | Release date: | 2020-08-19 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-guided covalent stabilization of coronavirus spike glycoprotein trimers in the closed conformation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4W5K
 
 | |
5K9F
 
 | |
5JSC
 
 | |
5KF0
 
 | |
5KIA
 
 | |
5KHA
 
 | |
5KOI
 
 | |
5KWV
 
 | |
