3C6E
 
 | |
1C47
 
 | |
1C4G
 
 | |
6UZC
 
 | |
7KH1
 
 | | Baseplate Complex for Myoviridae Phage XM1 | | Descriptor: | baseplate organization protein, gp11, baseplate stabilizing protein, ... | | Authors: | Wang, Z, Klose, T, Jiang, W, Kuhn, R.J. | | Deposit date: | 2020-10-19 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of Vibrio phage XM1, a simple contractile DNA injection machine
Biorxiv, 2021
|
|
7KLN
 
 | | Myoviridae Phage XM1 Neck Region (12-fold) | | Descriptor: | Head completion protein, gp1, Portal protein | | Authors: | Wang, Z, Klose, T, Jiang, W, Kuhn, R.J. | | Deposit date: | 2020-10-30 | | Release date: | 2021-11-03 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of Vibrio phage XM1, a simple contractile DNA injection machine
Biorxiv, 2021
|
|
7KJK
 
 | | The Neck region of Phage XM1 (6-fold symmetry) | | Descriptor: | Collar spike protein, Head completion protein, Tail sheath protein, ... | | Authors: | Wang, Z, Klose, T, Jiang, W, Kuhn, R.J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of Vibrio phage XM1, a simple contractile DNA injection machine
Biorxiv, 2021
|
|
7KMX
 
 | | The capsid of Myoviridae Phage XM1 | | Descriptor: | Major capsid protein, Minor capsid protein | | Authors: | Wang, Z, Klose, T, Jiang, W, Kuhn, R.J. | | Deposit date: | 2020-11-03 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of Vibrio phage XM1, a simple contractile DNA injection machine
Biorxiv, 2021
|
|
7LCG
 
 | | The mature Usutu SAAR-1776, Model A | | Descriptor: | (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Khare, B, Klose, T, Fang, Q, Kuhn, R. | | Deposit date: | 2021-01-11 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structure of Usutu virus SAAR-1776 displays fusion loop asymmetry.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LCH
 
 | | The mature Usutu SAAR-1776, Model B | | Descriptor: | (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Khare, B, Klose, T, Fang, Q, Kuhn, R. | | Deposit date: | 2021-01-11 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structure of Usutu virus SAAR-1776 displays fusion loop asymmetry.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N69
 
 | |
7N6A
 
 | |
1ZKU
 
 | |
1P5Y
 
 | |
1P5W
 
 | |
1HRI
 
 | | STRUCTURE DETERMINATION OF ANTIVIRAL COMPOUND SCH 38057 COMPLEXED WITH HUMAN RHINOVIRUS 14 | | Descriptor: | 1-[6-(2-CHLORO-4-METHYXYPHENOXY)-HEXYL]-IMIDAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Zhang, A, Nanni, R.G, Oren, D.A, Arnold, E. | | Deposit date: | 1992-10-01 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure determination of antiviral compound SCH 38057 complexed with human rhinovirus 14.
J.Mol.Biol., 230, 1993
|
|
1HRV
 
 | | HRV14/SDZ 35-682 COMPLEX | | Descriptor: | 1-[2-HYDROXY-3-(4-CYCLOHEXYL-PHENOXY)-PROPYL]-4-(2-PYRIDYL)-PIPERAZINE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Oren, D.A, Zhang, A, Arnold, E. | | Deposit date: | 1995-03-02 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | SDZ 35-682, a new picornavirus capsid-binding agent with potent antiviral activity.
Antiviral Res., 26, 1995
|
|
2CAH
 
 | |
3PMG
 
 | |
2CAG
 
 | | CATALASE COMPOUND II | | Descriptor: | CATALASE COMPOUND II, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gouet, P, Jouve, H.M, Hajdu, J. | | Deposit date: | 1996-06-12 | | Release date: | 1996-12-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ferryl intermediates of catalase captured by time-resolved Weissenberg crystallography and UV-VIS spectroscopy.
Nat.Struct.Biol., 3, 1996
|
|
3GK8
 
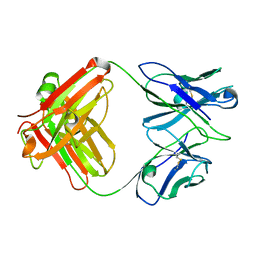 | | X-ray crystal structure of the Fab from MAb 14, mouse antibody against Canine Parvovirus | | Descriptor: | Fab 14 Heavy Chain, Fab 14 Light Chain | | Authors: | Hafenstein, S, Bowman, V, Sun, T, Nelson, C, Palermo, L, Chipman, P, Battisti, A, Parrish, C. | | Deposit date: | 2009-03-10 | | Release date: | 2009-06-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids.
J.Virol., 83, 2009
|
|
1CF9
 
 | |
3I50
 
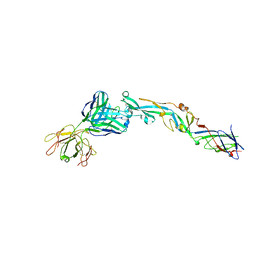 | | Crystal structure of the West Nile Virus envelope glycoprotein in complex with the E53 antibody Fab | | Descriptor: | Envelope glycoprotein, murine heavy chain (IgG3) of E53 monoclonal antibody Fab, murine kappa light chain of E53 monoclonal antibody Fab | | Authors: | Nybakken, G.E, Warren, J.T, Chen, B.R, Nelson, C.A, Fremont, D.H. | | Deposit date: | 2009-07-03 | | Release date: | 2009-10-27 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody.
Embo J., 28, 2009
|
|
1VKL
 
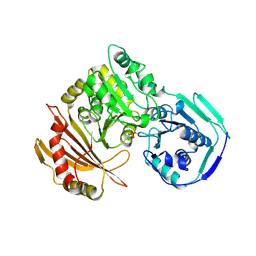 | | RABBIT MUSCLE PHOSPHOGLUCOMUTASE | | Descriptor: | NICKEL (II) ION, PHOSPHOGLUCOMUTASE | | Authors: | Ray Junior, W.J, Baranidharan, S, Liu, Y. | | Deposit date: | 1996-07-03 | | Release date: | 1997-01-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural changes at the metal ion binding site during the phosphoglucomutase reaction.
Biochemistry, 32, 1993
|
|
1E93
 
 | | High resolution structure and biochemical properties of a recombinant catalase depleted in iron | | Descriptor: | ACETATE ION, CATALASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Andreoletti, P, Sainz, G, Jaquinod, M, Gagnon, J, Jouve, H.M. | | Deposit date: | 2000-10-05 | | Release date: | 2000-10-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-Resolution Structure and Biochemical Properties of a Recombinant Proteus Mirabilis Catalase Depleted in Iron.
Proteins: Struct.,Funct., Genet., 50, 2003
|
|
