4R2W
 
 | | X-ray structure of uridine phosphorylase from Shewanella oneidensis MR-1 in complex with uridine at 1.6 A resolution | | 分子名称: | GLYCEROL, SULFATE ION, URIDINE, ... | | 著者 | Safonova, T.N, Mordkovich, N.N, Manuvera, V.A, Veiko, V.P, Popov, V.O, Polyakov, K.P. | | 登録日 | 2014-08-13 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | High-syn conformation of uridine and asymmetry of the hexameric molecule revealed in the high-resolution structures of Shewanella oneidensis MR-1 uridine phosphorylase in the free form and in complex with uridine.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4R2X
 
 | | Unique conformation of uridine and asymmetry of the hexameric molecule revealed in the high-resolution structures of Shewanella oneidensis uridine phosphorylase in the free form and in complex with uridine | | 分子名称: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | 著者 | Safonova, T.N, Mordkovich, N.N, Manuvera, V.A, Veiko, V.P, Popov, V.O, Polyakov, K.M. | | 登録日 | 2014-08-13 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (0.93 Å) | | 主引用文献 | High-syn conformation of uridine and asymmetry of the hexameric molecule revealed in the high-resolution structures of Shewanella oneidensis MR-1 uridine phosphorylase in the free form and in complex with uridine.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7OIN
 
 | | Crystal structure of LSSmScarlet - a genetically encoded red fluorescent protein with a large Stokes shift | | 分子名称: | LSSmScarlet - Genetically Encoded Red Fluorescent Proteins with a Large Stokes Shift, SODIUM ION, SULFATE ION | | 著者 | Boyko, K.M, Nikolaeva, A.Y, Dorovatovskii, P.V, Subach, O.M, Vlaskina, A.V, Agapova, Y.K, Ivashkina, O.I, Popov, V.O, Subach, F.V. | | 登録日 | 2021-05-12 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | LSSmScarlet, dCyRFP2s, dCyOFP2s and CRISPRed2s, Genetically Encoded Red Fluorescent Proteins with a Large Stokes Shift.
Int J Mol Sci, 22, 2021
|
|
7O45
 
 | | Crystal structure of ADD domain of the human DNMT3B methyltransferase | | 分子名称: | BROMIDE ION, Isoform 6 of DNA (cytosine-5)-methyltransferase 3B, ZINC ION | | 著者 | Boyko, K.M, Nikolaeva, A.Y, Bonchuk, A.N, Georgiev, P.G, Popov, V.O. | | 登録日 | 2021-04-05 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the DNMT3B ADD domain suggests the absence of a DNMT3A-like autoinhibitory mechanism.
Biochem.Biophys.Res.Commun., 619, 2022
|
|
7P7X
 
 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis (holo form). | | 分子名称: | ACETATE ION, Aminotransferase class IV, PHOSPHATE ION, ... | | 著者 | Boyko, K.M, Nikolaeva, A.Y, Bakunova, A.K, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O. | | 登録日 | 2021-07-20 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Uncommon Active Site of D-Amino Acid Transaminase from Haliscomenobacter hydrossis : Biochemical and Structural Insights into the New Enzyme.
Molecules, 26, 2021
|
|
7P8O
 
 | | Crystal structure of D-aminoacid transaminase from Haliscomenobacter hydrossis in its intermediate form | | 分子名称: | Aminotransferase class IV, MAGNESIUM ION, SULFATE ION | | 著者 | Matyuta, I.O, Boyko, K.M, Bakunova, A.K, Nikolaeva, A.Y, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O. | | 登録日 | 2021-07-23 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of D-aminoacid transaminase from Haliscomenobacter hydrossis in its apo form
To Be Published
|
|
7PPP
 
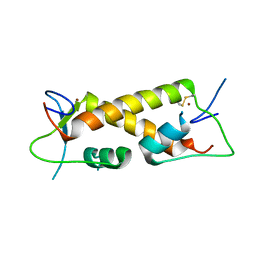 | | Crystal structure of ZAD-domain of ZNF_276 protein from rabbit. | | 分子名称: | ZINC ION, Zinc finger protein 276 | | 著者 | Boyko, K.M, Bonchuk, A.N, Nikolaeva, A.Y, Georgiev, P.G, Popov, V.O. | | 登録日 | 2021-09-14 | | 公開日 | 2021-12-08 | | 最終更新日 | 2022-07-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure, 30, 2022
|
|
7POK
 
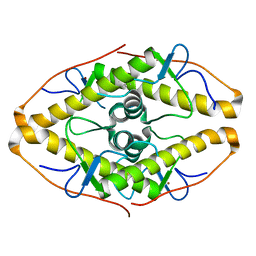 | | Crystal structure of ZAD-domain of Pita protein from D.melanogaster | | 分子名称: | LD15650p, ZINC ION | | 著者 | Boyko, K.M, Bonchuk, A.N, Nikolaeva, A.Y, Georgiev, P.G, Popov, V.O. | | 登録日 | 2021-09-09 | | 公開日 | 2021-12-08 | | 最終更新日 | 2022-07-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure, 30, 2022
|
|
7PO9
 
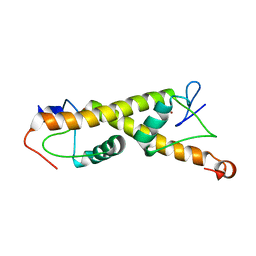 | | Crystal structure of ZAD-domain of M1BP protein from D.melanogaster | | 分子名称: | LD30467p, ZINC ION | | 著者 | Boyko, K.M, Bonchuk, A.N, Nikolaeva, A.Y, Georgiev, P.G, Popov, V.O. | | 登録日 | 2021-09-08 | | 公開日 | 2021-12-08 | | 最終更新日 | 2022-07-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure, 30, 2022
|
|
7POH
 
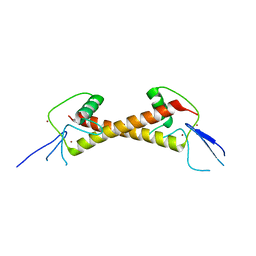 | | Crystal structure of ZAD-domain of Serendipity-d protein from D.melanogaster | | 分子名称: | Serendipity locus protein delta, ZINC ION | | 著者 | Boyko, K.M, Kachalova, G.S, Bonchuk, A.N, Nikolaeva, A.Y, Georgiev, P.G, Popov, V.O. | | 登録日 | 2021-09-09 | | 公開日 | 2021-12-08 | | 最終更新日 | 2022-07-20 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure, 30, 2022
|
|
7Q6B
 
 | | mRubyFT/S148I, a mutant of blue-to-red fluorescent timer in its blue state | | 分子名称: | mRubyFT S148I, a mutant of blue-to-red fluorescent timer | | 著者 | Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Dorovatovskii, P.V, Khrenova, M.G, Subach, O.M, Popov, V.O, Subach, F.M. | | 登録日 | 2021-11-06 | | 公開日 | 2023-04-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Combined Structural and Computational Study of the mRubyFT Fluorescent Timer Locked in Its Blue Form.
Int J Mol Sci, 24, 2023
|
|
7QGK
 
 | | The mRubyFT protein, Genetically Encoded Blue-to-Red Fluorescent Timer in its red state | | 分子名称: | MAGNESIUM ION, The red form of the mRubyFT protein, Genetically Encoded Blue-to-Red Fluorescent Timer | | 著者 | Boyko, K.M, Nikolaeva, A.Y, Gaivoronskii, F.A, Vlaskina, A.V, Subach, O.M, Popov, V.O, Subach, F.V. | | 登録日 | 2021-12-08 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The mRubyFT Protein, Genetically Encoded Blue-to-Red Fluorescent Timer.
Int J Mol Sci, 23, 2022
|
|
8ONJ
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant R88L | | 分子名称: | Aminotransferase class IV, DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | 登録日 | 2023-04-03 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
8ONL
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A | | 分子名称: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | 登録日 | 2023-04-03 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
8ONN
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A complexed with 3-aminooxypropionic acid | | 分子名称: | 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV | | 著者 | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | 登録日 | 2023-04-03 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
8PEI
 
 | | Crystal structure of the biphotochromic fluorescent protein SAASoti (C21N/V127T variant) in its green on-state | | 分子名称: | C21N/V127T form of the biphotochromic fluorescent protein SAASoti | | 著者 | Boyko, K.M, Varfolomeeva, L.A, Matyuta, I.O, Gavshina, A.V, Solovyev, I.D, Popov, V.O, Savitsky, A.P. | | 登録日 | 2023-06-14 | | 公開日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of the biphotochromic fluorescent protein C21N/V127T SAASoti in its green on-state
To Be Published
|
|
8ONM
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A complexed with D-glutamate | | 分子名称: | (~{Z})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]pent-2-enedioic acid, 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | 登録日 | 2023-04-03 | | 公開日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Probing of the structural and catalytic roles of the residues in the active site of transaminase from Aminobacterium colombiense
To Be Published
|
|
8PYH
 
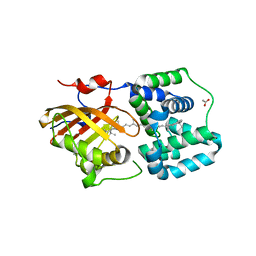 | | Crystal structure of the Orange Carotenoid Protein 2 (OCP2) from Crinalium epipsammum PCC 9333 | | 分子名称: | ACETATE ION, Orange carotenoid-binding protein, beta,beta-caroten-4-one | | 著者 | Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Maksimov, E.G, Popov, V.O, Sluchanko, N.N. | | 登録日 | 2023-07-25 | | 公開日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural framework for the understanding spectroscopic and functional signatures of the cyanobacterial Orange Carotenoid Protein families.
Int.J.Biol.Macromol., 254, 2024
|
|
8Q9X
 
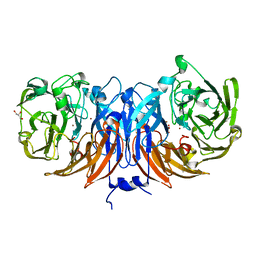 | | The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum with molecular oxygen at 1.05 A resolution | | 分子名称: | COPPER (II) ION, GLYCEROL, OXYGEN MOLECULE, ... | | 著者 | Varfolomeeva, L.A, Polyakov, K.M, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | 登録日 | 2023-08-22 | | 公開日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum at atomic resolution
To Be Published
|
|
8Q9Y
 
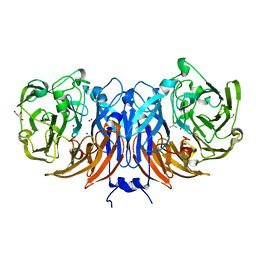 | | The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum in complex with inhibitor thiourea at 1.10 A resolution | | 分子名称: | COPPER (II) ION, GLYCEROL, THIOUREA, ... | | 著者 | Varfolomeeva, L.A, Polyakov, K.M, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | 登録日 | 2023-08-22 | | 公開日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum at atomic resolution
To Be Published
|
|
8PZK
 
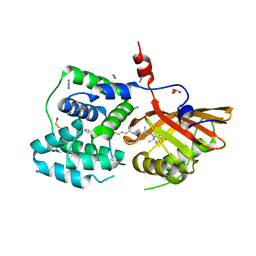 | | Crystal structure of the Orange Carotenoid Protein 2 (OCP2) from Gloeocapsa sp. PCC 7428 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, GLYCEROL, ... | | 著者 | Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Maksimov, E.G, Popov, V.O, Sluchanko, N.N. | | 登録日 | 2023-07-27 | | 公開日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural framework for the understanding spectroscopic and functional signatures of the cyanobacterial Orange Carotenoid Protein families.
Int.J.Biol.Macromol., 254, 2024
|
|
8RAI
 
 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis point mutant R90I complexed with phenylhydrazine | | 分子名称: | Aminotransferase class IV, GLYCEROL, [6-methyl-5-oxidanyl-4-[(2-phenylhydrazinyl)methyl]pyridin-3-yl]methyl dihydrogen phosphate | | 著者 | Matyuta, I.O, Bakunova, A.K, Minyaev, M.E, Popov, V.O, Bezsudnova, E.Y, Boyko, K.M. | | 登録日 | 2023-12-01 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Multifunctionality of arginine residues in the active sites of non-canonical d-amino acid transaminases.
Arch.Biochem.Biophys., 756, 2024
|
|
8RAF
 
 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis point mutant R90I (holo form) | | 分子名称: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Matyuta, I.O, Bakunova, A.K, Minyaev, M.E, Popov, V.O, Bezsudnova, E.Y, Boyko, K.M. | | 登録日 | 2023-12-01 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Multifunctionality of arginine residues in the active sites of non-canonical d-amino acid transaminases.
Arch.Biochem.Biophys., 756, 2024
|
|
3TTB
 
 | | Structure of the Thioalkalivibrio paradoxus cytochrome c nitrite reductase in complex with sulfite | | 分子名称: | CALCIUM ION, COBALT (II) ION, Eight-heme nitrite reductase, ... | | 著者 | Polyakov, K.M, Trofimov, A.A, Tikhonova, T.V, Tikhonov, A.V, Dorovatovskii, P.V, Popov, V.O. | | 登録日 | 2011-09-14 | | 公開日 | 2011-10-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Comparative structural and functional analysis of two octaheme nitrite reductases from closely related Thioalkalivibrio species.
Febs J., 279, 2012
|
|
3V9E
 
 | | Structure of the L499M mutant of the laccase from B.aclada | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, GLYCEROL, ... | | 著者 | Osipov, E.M, Polyakov, K.M, Tikhonova, T.V, Dorovatovsky, P.V, Ludwig, R, Kittl, R, Shleev, S.V, Popov, V.O. | | 登録日 | 2011-12-27 | | 公開日 | 2013-01-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Effect of the L499M mutation of the ascomycetous Botrytis aclada laccase on redox potential and catalytic properties.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
