4RGZ
 
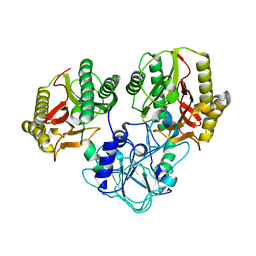 | | Crystal structure of recombinant prolidase from Thermococcus sibiricus at P21221 spacegroup | | Descriptor: | PHOSPHATE ION, Xaa-Pro aminopeptidase, ZINC ION | | Authors: | Timofeev, V.I, Korgenevsky, D.A, Gorbacheva, M.A, Boyko, K.M, Slutsky, E, Rakitina, T.V, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2014-10-01 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of recombinant prolidase from Thermococcus sibiricus in space group P21221.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
2OGR
 
 | |
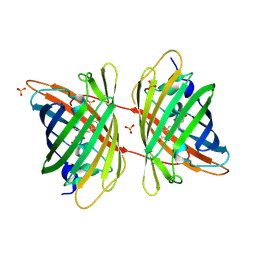
3GB3
 
 | |
3SXQ
 
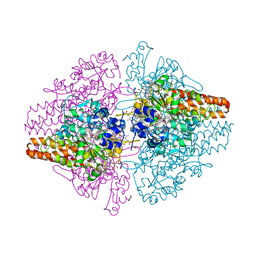 | | Structure of a hexameric multiheme c nitrite reductase from the extremophile bacterium Thiolkalivibrio paradoxus | | Descriptor: | CALCIUM ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Polyakov, K.M, Trofimov, A.A, Tikhonova, T.V, Tikhonov, A.V, Boyko, K.M, Popov, V.O. | | Deposit date: | 2011-07-15 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparative structural and functional analysis of two octaheme nitrite reductases from closely related Thioalkalivibrio species.
Febs J., 279, 2012
|
|
3SQR
 
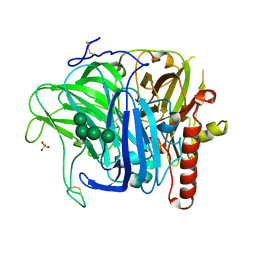 | | Crystal structure of laccase from Botrytis aclada at 1.67 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, SULFATE ION, ... | | Authors: | Osipov, E.M, Polyakov, K.M, Tikhonova, T.V, Dorovatovsky, P.V, Ludwig, R, Kittl, R, Shleev, S.V, Popov, V.O. | | Deposit date: | 2011-07-06 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Effect of the L499M mutation of the ascomycetous Botrytis aclada laccase on redox potential and catalytic properties.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8OQ0
 
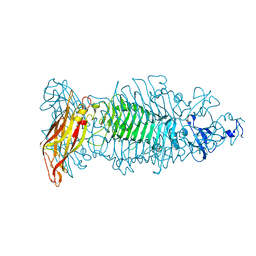 | | Crystal structure of tailspike depolymerase (APK09_gp48) from Acinetobacter phage APK09 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tailspike protein | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shneider, M.M, Timoshina, O.Y, Popova, A.V, Miroshnikov, K.A, Popov, V.O. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Friunavirus Phage-Encoded Depolymerases Specific to Different Capsular Types of Acinetobacter baumannii .
Int J Mol Sci, 24, 2023
|
|
6FP5
 
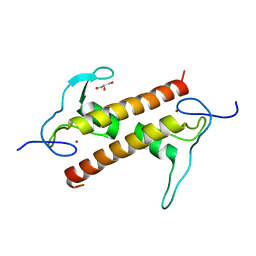 | | Crystal structure of ZAD-domain of CG2712 protein from D.melanogaster | | Descriptor: | CG2712, GLYCEROL, ZINC ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Bonchuk, A.N, Kachalova, G.S, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2018-02-09 | | Release date: | 2019-08-21 | | Last modified: | 2021-04-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of diversity and homodimerization specificity of zinc-finger-associated domains in Drosophila.
Nucleic Acids Res., 49, 2021
|
|
8C0N
 
 | | Crystal structure of the red form of the mTagFT fluorescent timer | | Descriptor: | Blue-to-red TagFT fluorescent timer | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Agapova, Y.K, Subach, O.M, Popov, V.O, Subach, F.V. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Blue-to-Red TagFT, mTagFT, mTsFT, and Green-to-FarRed mNeptusFT2 Proteins, Genetically Encoded True and Tandem Fluorescent Timers.
Int J Mol Sci, 24, 2023
|
|
6TTB
 
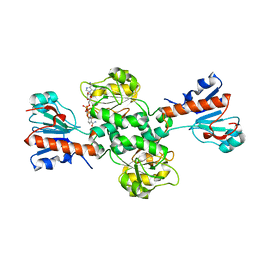 | | Crystal structure of NAD-dependent formate dehydrogenase from Staphylococcus aureus in complex with NAD | | Descriptor: | Formate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Boyko, K.M, Pometun, A.A, Nikolaeva, A.Y, Kargov, I.S, Yurchenko, T.S, Savin, S.S, Popov, V.O, Tishkov, V.I. | | Deposit date: | 2019-12-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of NAD-dependent formate dehydrogenase from Staphylococcus aureus in complex with NAD
To Be Published
|
|
8OQ1
 
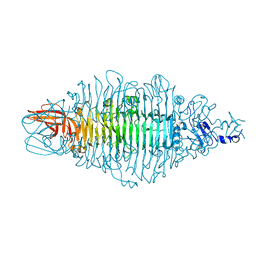 | | Crystal structure of tailspike depolymerase (APK14_gp49) from Acinetobacter phage vB_AbaP_APK14 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shneider, M.M, Timoshina, O.Y, Miroshnikov, K.A, Popov, V.O. | | Deposit date: | 2023-04-10 | | Release date: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Friunavirus Phage-Encoded Depolymerases Specific to Different Capsular Types of Acinetobacter baumannii.
Int J Mol Sci, 24, 2023
|
|
5MR0
 
 | | Thermophilic archaeal branched-chain amino acid transaminases from Geoglobus acetivorans and Archaeoglobus fulgidus: biochemical and structural characterisation | | Descriptor: | 1,2-ETHANEDIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, CHLORIDE ION, ... | | Authors: | Isupov, M.N, Littlechild, J.A, James, P, Sayer, C, Sutter, J.M, Schmidt, M, Schoenheit, P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the ArchaeaGeoglobus acetivoransandArchaeoglobus fulgidus: Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
5MQZ
 
 | | Archaeal branched-chain amino acid aminotransferase from Archaeoglobus fulgidus; holoform | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | James, P, Isupov, M.N, Sayer, C, Littlechild, J.A, Sutter, J.M, Schmidt, M, Schoenheit, P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus acetivorans and Archaeoglobus fulgidus : Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
8P1H
 
 | | Crystal structure of the chimera of human 14-3-3 zeta and phosphorylated cytoplasmic loop fragment of the alpha7 acetylcholine receptor | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, BENZOIC ACID, ... | | Authors: | Boyko, K.M, Kapitonova, A.A, Tugaeva, K.V, Varfolomeeva, L.A, Lyukmanova, E.N, Sluchanko, N.N. | | Deposit date: | 2023-05-12 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure reveals canonical recognition of the phosphorylated cytoplasmic loop of human alpha7 nicotinic acetylcholine receptor by 14-3-3 protein.
Biochem.Biophys.Res.Commun., 682, 2023
|
|
4ZBL
 
 | |
8OPZ
 
 | | Crystal structure of a tailspike depolymerase (APK16_gp47) from Acinetobacter phage APK16 | | Descriptor: | GLYCEROL, Tailspike depolymerase (APK16_gp47) from Acinetobacter phage APK16 | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shneider, M.M, Timoshina, O.Y, Miroshnikov, K.A, Popov, V.O. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Friunavirus Phage-Encoded Depolymerases Specific to Different Capsular Types of Acinetobacter baumannii .
Int J Mol Sci, 24, 2023
|
|
8AAB
 
 | | S148F mutant of blue-to-red fluorescent timer mRubyFT | | Descriptor: | mRubyFT S148F mutant of blue-to-red fluorescent timer | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Dorovatovskii, P.V, Subach, O.M, Popov, V.O, Subach, F.V. | | Deposit date: | 2022-06-30 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | mRubyFT/S147I, a mutant of blue-to-red fluorescent timer
Crystallography Reports, 2022
|
|
8P6G
 
 | | Crystal structure of the improved version of the Genetically Encoded Green Calcium Indicator YTnC2-5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Genetically Encoded Green Calcium Indicator YTnC2-5 based on Troponin C from toadfish | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Subach, O.M, Vlaskina, A.V, Agapova, Y.K, Popov, V.O, Subach, F.V. | | Deposit date: | 2023-05-26 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | YTnC2, an improved genetically encoded green calcium indicator based on toadfish troponin C.
Febs Open Bio, 13, 2023
|
|
8QND
 
 | | Crystal structure of the ribonucleoside hydrolase C from Lactobacillus reuteri | | Descriptor: | CALCIUM ION, Inosine-uridine nucleoside N-ribohydrolase | | Authors: | Matyuta, I.O, Minyaev, M.E, Shaposhnikov, L.A, Pometun, E.V, Tishkov, V.I, Popov, V.O, Boyko, K.M. | | Deposit date: | 2023-09-26 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Functional Examination of Novel Ribonucleoside Hydrolase C (RihC) from Limosilactobacillus reuteri LR1.
Int J Mol Sci, 25, 2023
|
|
