5LMM
 
 | | Structure of E coli Hydrogenase Hyd-1 mutant E28Q | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, ... | | Authors: | Carr, S.B, Phillips, S.E.V, Evans, R.M, Brooke, E.J, Armstrong, F.A. | | Deposit date: | 2016-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Kinetic consequences of re-engineering the outer shell "canopy" above the active site of a [NiFe]-hydrogenase.
To Be Published
|
|
5MVL
 
 | | Crystal structure of an A-DNA dodecamer containing 5-bromouracil | | Descriptor: | Brominated DNA dodecamer, MAGNESIUM ION | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-16 | | Release date: | 2017-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.405 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVT
 
 | | Crystal structure of an A-DNA dodecamer featuring an alternating pyrimidine-purine sequence | | Descriptor: | COBALT (III) ION, DNA | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVQ
 
 | | Crystal structure of an unmodified, self-complementary dodecamer. | | Descriptor: | DNA, MAGNESIUM ION | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVU
 
 | |
5MVP
 
 | | Crystal structure of an A-DNA dodecamer containing the GGGCCC motif | | Descriptor: | DNA, POTASSIUM ION | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVK
 
 | |
1GOH
 
 | |
1GOG
 
 | |
1GOF
 
 | | NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE | | Descriptor: | ACETIC ACID, COPPER (II) ION, GALACTOSE OXIDASE, ... | | Authors: | Ito, N, Phillips, S.E.V, Knowles, P.F. | | Deposit date: | 1993-09-30 | | Release date: | 1994-01-31 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel thioether bond revealed by a 1.7 A crystal structure of galactose oxidase.
Nature, 350, 1991
|
|
1BQ4
 
 | |
2JKX
 
 | | Galactose oxidase. MatGO. Copper free, expressed in Pichia Pastoris. | | Descriptor: | ACETATE ION, CALCIUM ION, GALACTOSE OXIDASE | | Authors: | Deacon, S.E, Mahmoud, K, Spooner, R.K, Firbank, S.J, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2008-09-01 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Enhanced Fructose Oxidase Activity in a Galactose Oxidase Variant
Chembiochem, 5, 2004
|
|
2LZ2
 
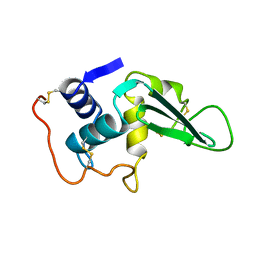 | |
1YP7
 
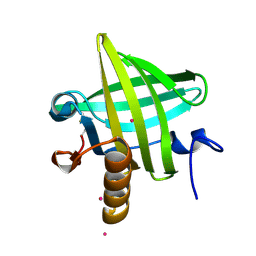 | | Van der Waals Interactions Dominate Hydrophobic Association in a Protein Binding Site Occluded From Solvent Water | | Descriptor: | CADMIUM ION, MAJOR URINARY PROTEIN 1 | | Authors: | Barratt, E, Bingham, R.J, Warner, D.J, Laughton, C.A, Phillips, S.E.V, Homans, S.W. | | Deposit date: | 2005-01-30 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Van der Waals Interactions Dominate Ligand-Protein Association in a Protein Binding Site Occluded from Solvent Water
J.Am.Chem.Soc., 127, 2005
|
|
1YP6
 
 | | Van der Waals Interactions Dominate Hydrophobic Association in a Protein Binding Site Occluded From Solvent Water | | Descriptor: | 2-ISOBUTYL-3-METHOXYPYRAZINE, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Barratt, E, Bingham, R.J, Warner, D.J, Laughton, C.A, Phillips, S.E.V, Homans, S.W. | | Deposit date: | 2005-01-30 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Van der Waals Interactions Dominate Ligand-Protein Association in a Protein Binding Site Occluded from Solvent Water
J.Am.Chem.Soc., 127, 2005
|
|
1ZSE
 
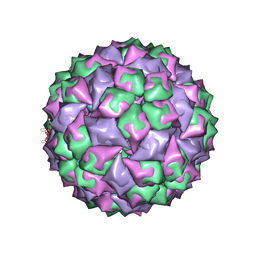 | | RNA stemloop from bacteriophage Qbeta complexed with an N87S mutant MS2 Capsid | | Descriptor: | Coat protein, RNA HAIRPIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-05-24 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2
Structure, 14, 2006
|
|
2B2D
 
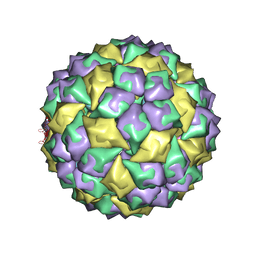 | | RNA stemloop operator from bacteriophage QBETA complexed with an N87S,E89K mutant MS2 capsid | | Descriptor: | 5'-R(*AP*UP*GP*CP*AP*UP*GP*UP*CP*UP*AP*AP*GP*AP*CP*AP*GP*CP*AP*U)-3', Coat protein | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-09-19 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of RNA binding discrimination between bacteriophages Qbeta and MS2
Structure, 14, 2006
|
|
2B2G
 
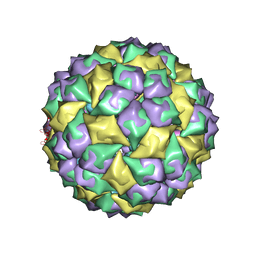 | | MS2 Wild-type RNA stemloop complexed with an N87S mutant MS2 capsid | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP*UP*AP*CP*CP*CP*AP*UP*GP*U)-3', Coat protein | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-09-19 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural basis of RNA binding discrimination between bacteriophages Qbeta and MS2
Structure, 14, 2006
|
|
2B2E
 
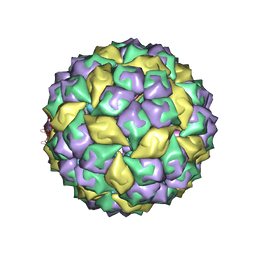 | | RNA stemloop from bacteriophage MS2 complexed with an N87S,E89K mutant MS2 capsid | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP*UP*AP*CP*CP*CP*AP*UP*GP*U)-3', Coat protein | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-09-19 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of RNA binding discrimination between bacteriophages Qbeta and MS2
Structure, 14, 2006
|
|
2BS0
 
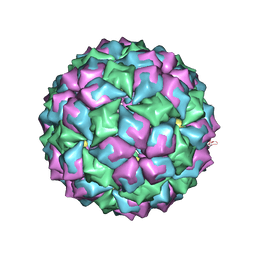 | | MS2 (N87AE89K mutant) - Variant Qbeta RNA hairpin complex | | Descriptor: | 5'-R(*AP*UP*GP*CP*AP*UP*GP*UP*CP*UP *AP*AP*GP*AP*CP*UP*GP*CP*AP*U)-3', COAT PROTEIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-05-13 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2.
Structure, 14, 2006
|
|
2BNY
 
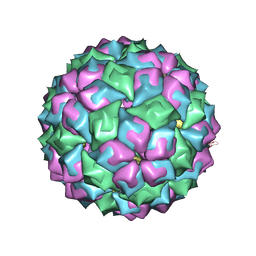 | | MS2 (N87A mutant) - RNA hairpin complex | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *UP*AP*CP*CP*CP*AP*UP*GP*U)-3', MS2 COAT PROTEIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-04-06 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2.
Structure, 14, 2006
|
|
2BQ5
 
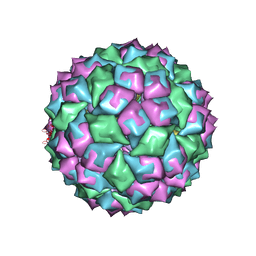 | | MS2 (N87AE89K mutant) - RNA hairpin complex | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *UP*AP*CP*CP*CP*AP*UP*GP*U)-3', COAT PROTEIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-04-27 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2.
Structure, 14, 2006
|
|
2BS1
 
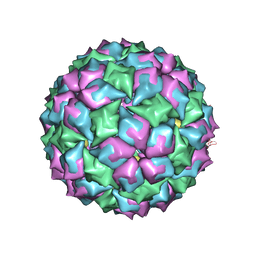 | | MS2 (N87AE89K mutant) - Qbeta RNA hairpin complex | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *UP*AP*CP*CP*CP*AP*UP*GP*U)-3', MS2 COAT PROTEIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-05-13 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2.
Structure, 14, 2006
|
|
1DYU
 
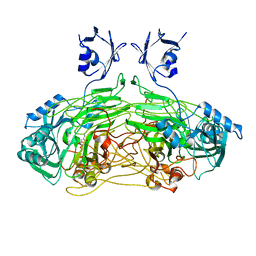 | | The active site base controls cofactor reactivity in Escherichia coli amine oxidase: X-ray crystallographic studies with mutational variants. | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE | | Authors: | Murray, J.M, Wilmot, C.M, Saysell, C.G, Jaeger, J, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2000-02-08 | | Release date: | 2000-02-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Active Site Base Controls Cofactor Reactivity in Escherichia Coli Amine Oxidase : X-Ray Crystallographicstudies with Mutational Variants
Biochemistry, 38, 1999
|
|
1CMA
 
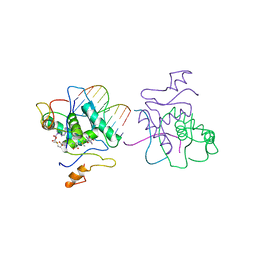 | | MET REPRESSOR/DNA COMPLEX + S-ADENOSYL-METHIONINE | | Descriptor: | DNA (5'-D(*AP*GP*AP*CP*GP*TP*CP*TP*A)-3'), DNA (5'-D(*TP*TP*AP*GP*AP*CP*GP*TP*CP*T)-3'), PROTEIN (MET REPRESSOR), ... | | Authors: | Somers, W.S, Phillips, S.E.V. | | Deposit date: | 1992-08-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands.
Nature, 359, 1992
|
|
