6OEZ
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor (+)-N-(Cyclobutylmethyl)-3-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-2-(1-{[(2S)-pyrro-lidin-2-yl]methyl}-1H-indol-5-yl)-1,3-thiazol-4-yl}prop-2-yn-1-amine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, N-(cyclobutylmethyl)-3-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-2-(1-{[(2S)-pyrrolidin-2-yl]methyl}-1H-indol-5-yl)-1,3-thiazol-4-yl}prop-2-yn-1-amine, ... | | Authors: | Halgas, O, De Gasparo, R, Harangozo, D, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting a Large Active Site: Structure-Based Design of Nanomolar Inhibitors of Trypanosoma brucei Trypanothione Reductase.
Chemistry, 25, 2019
|
|
6QKU
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Tyr219Phe - Chloroacetate soaked 2hr | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, GLYCOLIC ACID, ... | | Authors: | Mehrabi, P, Kim, T.H, Prosser, R.S, Pai, E.F. | | Deposit date: | 2019-01-30 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | Substrate-Based Allosteric Regulation of a Homodimeric Enzyme.
J.Am.Chem.Soc., 141, 2019
|
|
5SWN
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Fluoroacetate - Cocrystallized | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5T4T
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn - Apo No Halide | | Descriptor: | ACETATE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-08-30 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
1NDA
 
 | | THE STRUCTURE OF TRYPANOSOMA CRUZI TRYPANOTHIONE REDUCTASE IN THE OXIDIZED AND NADPH REDUCED STATE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, TRYPANOTHIONE OXIDOREDUCTASE | | Authors: | Lantwin, C.B, Kabsch, W, Pai, E.F, Schlichting, I, Krauth-Siegel, R.L. | | Deposit date: | 1993-07-02 | | Release date: | 1994-09-30 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of Trypanosoma cruzi trypanothione reductase in the oxidized and NADPH reduced state.
Proteins, 18, 1994
|
|
1PLK
 
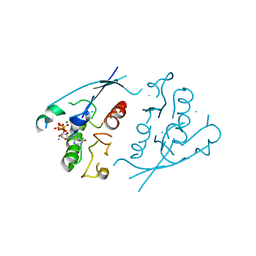 | | CRYSTALLOGRAPHIC STUDIES ON P21H-RAS USING SYNCHROTRON LAUE METHOD: IMPROVEMENT OF CRYSTAL QUALITY AND MONITORING OF THE GTPASE REACTION AT DIFFERENT TIME POINTS | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Scheidig, A, Sanchez-Llorente, A, Lautwein, A, Pai, E.F, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Goody, R.S. | | Deposit date: | 1994-03-13 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic studies on p21(H-ras) using the synchrotron Laue method: improvement of crystal quality and monitoring of the GTPase reaction at different time points.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1PLJ
 
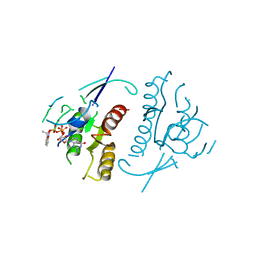 | | CRYSTALLOGRAPHIC STUDIES ON P21H-RAS USING SYNCHROTRON LAUE METHOD: IMPROVEMENT OF CRYSTAL QUALITY AND MONITORING OF THE GTPASE REACTION AT DIFFERENT TIME POINTS | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE 5'-TRIPHOSPHATE P3-[1-(2-NITROPHENYL)ETHYL ESTER], MAGNESIUM ION | | Authors: | Scheidig, A, Sanchez-Llorente, A, Lautwein, A, Pai, E.F, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Goody, R.S. | | Deposit date: | 1994-03-13 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic studies on p21(H-ras) using the synchrotron Laue method: improvement of crystal quality and monitoring of the GTPase reaction at different time points.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1PLL
 
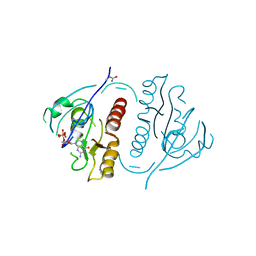 | | CRYSTALLOGRAPHIC STUDIES ON P21H-RAS USING SYNCHROTRON LAUE METHOD: IMPROVEMENT OF CRYSTAL QUALITY AND MONITORING OF THE GTPASE REACTION AT DIFFERENT TIME POINTS | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Scheidig, A, Sanchez-Llorente, A, Lautwein, A, Pai, E.F, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Goody, R.S. | | Deposit date: | 1994-03-13 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic studies on p21(H-ras) using the synchrotron Laue method: improvement of crystal quality and monitoring of the GTPase reaction at different time points.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
4XZJ
 
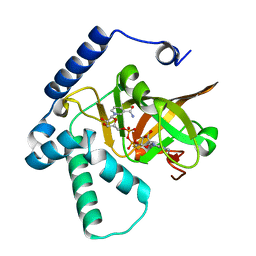 | | Crystal structure of ADP-ribosyltransferase Vis in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative NAD(+)--arginine ADP-ribosyltransferase Vis | | Authors: | Pfoh, R, Ravulapalli, R, Merrill, A.R, Pai, E.F. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of Vis Toxin, a Novel ADP-Ribosyltransferase from Vibrio splendidus.
Biochemistry, 54, 2015
|
|
4XZK
 
 | |
4YTY
 
 | | Structure of rat xanthine oxidoreductase, C535A/C992R/C1324S, NADH bound form | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Matsumura, T, Eger, B.T, Pai, E.F. | | Deposit date: | 2015-03-18 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase.
Febs J., 282, 2015
|
|
4YP5
 
 | | Crystal structure of Methanobacterium thermoautotrophicum NMNAT in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Pfoh, R, Christendat, D, Pai, E.F, Saridakis, V. | | Deposit date: | 2015-03-12 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Nicotinamide mononucleotide adenylyltransferase displays alternate binding modes for nicotinamide nucleotides.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4YSW
 
 | | Structure of rat xanthine oxidoreductase, C-terminal deletion protein variant, NADH bound form | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Matsumura, T, Eger, B.T, Pai, E.F. | | Deposit date: | 2015-03-17 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase.
Febs J., 282, 2015
|
|
4YTZ
 
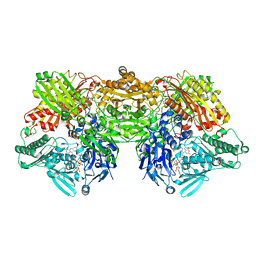 | | Rat xanthine oxidoreductase, C-terminal deletion protein variant, crystal grown without dithiothreitol | | Descriptor: | BICARBONATE ION, CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Matsumura, T, Eger, B.T, Pai, E.F, Nishino, T. | | Deposit date: | 2015-03-18 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase
Febs J., 282, 2015
|
|
4YP7
 
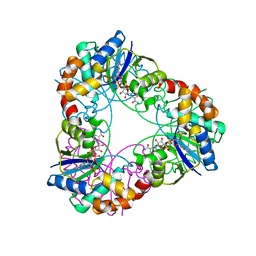 | | Crystal structure of Methanobacterium thermoautotrophicum NMNAT in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Pfoh, R, Christendat, D, Pai, E.F, Saridakis, V. | | Deposit date: | 2015-03-12 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nicotinamide mononucleotide adenylyltransferase displays alternate binding modes for nicotinamide nucleotides.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4YP6
 
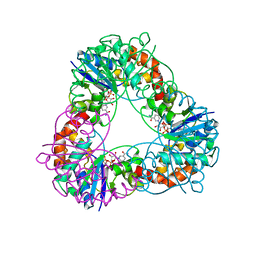 | | Crystal structure of Methanobacterium thermoautotrophicum NMNAT in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Pfoh, R, Christendat, D, Pai, E.F, Saridakis, V. | | Deposit date: | 2015-03-12 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nicotinamide mononucleotide adenylyltransferase displays alternate binding modes for nicotinamide nucleotides.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
1GNP
 
 | | X-RAY CRYSTAL STRUCTURE ANALYSIS OF THE CATALYTIC DOMAIN OF THE ONCOGENE PRODUCT P21H-RAS COMPLEXED WITH CAGED GTP AND MANT DGPPNHP | | Descriptor: | C-H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID 3'-O-(N-METHYLANTHRANILOYL-2'-DEOXYGUANYLATE ESTER | | Authors: | Scheidig, A, Franken, S.M, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Pai, E.F, Goody, R.S. | | Deposit date: | 1995-05-11 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray crystal structure analysis of the catalytic domain of the oncogene product p21H-ras complexed with caged GTP and mant dGppNHp.
J.Mol.Biol., 253, 1995
|
|
1YQE
 
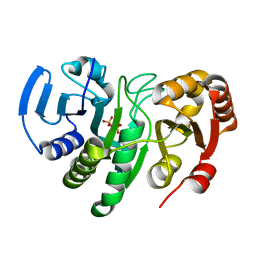 | | Crystal Structure of Conserved Protein of Unknown Function AF0625 | | Descriptor: | Hypothetical UPF0204 protein AF0625, PYROPHOSPHATE 2- | | Authors: | Liu, Y, Skarina, T, Dong, A, Kudritskam, M, Savchenko, A, Pai, E.F, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-01 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Conserved Hypothetical Protein AF0625
To be Published
|
|
6QHT
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 376 ms | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
6QI2
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 13536 ms | | Descriptor: | Fluoroacetate dehalogenase, GLYCOLIC ACID | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
6QHZ
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 6788 ms | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
6QKW
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Tyr219Phe - Fluoroacetate soaked 2hr | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, GLYCOLIC ACID, ... | | Authors: | Mehrabi, P, Kim, T.H, Prosser, R.S, Pai, E.F. | | Deposit date: | 2019-01-30 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.512 Å) | | Cite: | Substrate-Based Allosteric Regulation of a Homodimeric Enzyme.
J.Am.Chem.Soc., 141, 2019
|
|
6QHP
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 2256 ms covalent intermediate 1 | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
6QHX
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 6156 ms | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
6QHU
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 100 ms | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
