3BE9
 
 | | Structure-based design and synthesis of novel macrocyclic pyrazolo[1,5-a] [1,3,5]triazine compounds as potent inhibitors of protein kinase CK2 and their anticancer activities | | Descriptor: | 19-(cyclopropylamino)-4,6,7,15-tetrahydro-5H-16,1-(azenometheno)-10,14-(metheno)pyrazolo[4,3-o][1,3,9]triazacyclohexadecin-8(9H)-one, Casein kinase II subunit alpha | | Authors: | Nie, Z, Perretta, C, Erickson, P, Margosiak, S, Lu, J, Averill, A, Almassy, R, Chu, S. | | Deposit date: | 2007-11-16 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design and synthesis of novel macrocyclic pyrazolo[1,5-a] [1,3,5]triazine compounds as potent inhibitors of protein kinase CK2 and their anticancer activities.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2PVJ
 
 | | Structure-Based Design of Pyrazolo[1,5-a][1,3,5]triazine Derivatives as Potent Inhibitors of Protein Kinase CK2 | | Descriptor: | 2-(CYCLOHEXYLMETHYLAMINO)-4-(PHENYLAMINO)PYRAZOLO[1,5-A][1,3,5]TRIAZINE-8-CARBONITRILE, Casein kinase II subunit alpha | | Authors: | Nie, Z, Perretta, C, Erickson, P, Margosiak, S, Almassy, R, Lu, J, Averill, A, Yager, K.M, Chu, S. | | Deposit date: | 2007-05-09 | | Release date: | 2008-05-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design, synthesis, and study of pyrazolo[1,5-a][1,3,5]triazine derivatives as potent inhibitors of protein kinase CK2.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2PVL
 
 | | Structure-Based Design of Pyrazolo[1,5-a][1,3,5]triazine Derivatives as Potent Inhibitors of Protein Kinase CK2 | | Descriptor: | 2-(4-ETHYLPIPERAZIN-1-YL)-4-(PHENYLAMINO)PYRAZOLO[1,5-A][1,3,5]TRIAZINE-8-CARBONITRILE, Casein kinase II subunit alpha | | Authors: | Nie, Z, Perretta, C, Erickson, P, Margosiak, S, Almassy, R, Lu, J, Averill, A, Yager, K.M, Chu, S. | | Deposit date: | 2007-05-09 | | Release date: | 2008-05-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design, synthesis, and study of pyrazolo[1,5-a][1,3,5]triazine derivatives as potent inhibitors of protein kinase CK2.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2PVK
 
 | | Structure-Based Design of Pyrazolo[1,5-a][1,3,5]triazine Derivatives as Potent Inhibitors of Protein Kinase CK2 | | Descriptor: | 2-(4-CHLOROBENZYLAMINO)-4-(PHENYLAMINO)PYRAZOLO[1,5-A][1,3,5]TRIAZINE-8-CARBONITRILE, Casein kinase II subunit alpha | | Authors: | Nie, Z, Perretta, C, Erickson, P, Margosiak, S, Almassy, R, Lu, J, Averill, A, Yager, K.M, Chu, S. | | Deposit date: | 2007-05-09 | | Release date: | 2008-05-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design, synthesis, and study of pyrazolo[1,5-a][1,3,5]triazine derivatives as potent inhibitors of protein kinase CK2.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2PVH
 
 | | Structure-Based Design of Pyrazolo[1,5-a][1,3,5]triazine Derivatives as Potent Inhibitors of Protein Kinase CK2 | | Descriptor: | Casein kinase II subunit alpha, N,N'-DIPHENYLPYRAZOLO[1,5-A][1,3,5]TRIAZINE-2,4-DIAMINE | | Authors: | Nie, Z, Perretta, C, Erickson, P, Margosiak, S, Almassy, R, Lu, J, Averill, A, Yager, K.M, Chu, S. | | Deposit date: | 2007-05-09 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based design, synthesis, and study of pyrazolo[1,5-a][1,3,5]triazine derivatives as potent inhibitors of protein kinase CK2.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2PVN
 
 | | Structure-Based Design of Pyrazolo[1,5-a][1,3,5]triazine Derivatives as Potent Inhibitors of Protein Kinase CK2 | | Descriptor: | Casein kinase II subunit alpha, N-(3-(8-CYANO-4-(PHENYLAMINO)PYRAZOLO[1,5-A][1,3,5]TRIAZIN-2-YLAMINO)PHENYL)ACETAMIDE | | Authors: | Nie, Z, Perretta, C, Erickson, P, Margosiak, S, Almassy, R, Lu, J, Averill, A, Yager, K.M, Chu, S. | | Deposit date: | 2007-05-09 | | Release date: | 2008-05-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis, and study of pyrazolo[1,5-a][1,3,5]triazine derivatives as potent inhibitors of protein kinase CK2.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2PVM
 
 | | Structure-Based Design of Pyrazolo[1,5-a][1,3,5]triazine Derivatives as Potent Inhibitors of Protein Kinase CK2 | | Descriptor: | 4-(2-(1H-IMIDAZOL-4-YL)ETHYLAMINO)-2-(PHENYLAMINO)PYRAZOLO[1,5-A][1,3,5]TRIAZINE-8-CARBONITRILE, Casein kinase II subunit alpha | | Authors: | Nie, Z, Perretta, C, Erickson, P, Margosiak, S, Almassy, R, Lu, J, Averill, A, Yager, K.M, Chu, S. | | Deposit date: | 2007-05-09 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis, and study of pyrazolo[1,5-a][1,3,5]triazine derivatives as potent inhibitors of protein kinase CK2.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6CG1
 
 | | Crystal Structure of KDM4A with Compound 14 | | Descriptor: | 3-{[(4-fluorophenyl)methyl]amino}pyridine-4-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | Hosfield, D.J, Nie, Z. | | Deposit date: | 2018-02-19 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure-based design and discovery of potent and selective KDM5 inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6CG2
 
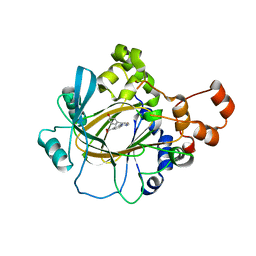 | | Crystal Structure of KDM4A with Compound 8 | | Descriptor: | 2-[5-(3-hydroxyphenyl)-1H-pyrazol-1-yl]pyridine-4-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | Hosfield, D.J, Nie, Z. | | Deposit date: | 2018-02-19 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure-based design and discovery of potent and selective KDM5 inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
3IL5
 
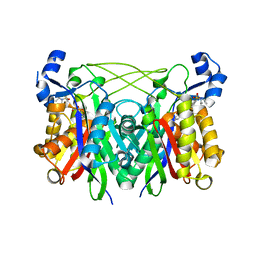 | | Structure of E. faecalis FabH in complex with 2-({4-bromo-3-[(diethylamino)sulfonyl]benzoyl}amino)benzoic acid | | Descriptor: | 2-({[4-bromo-3-(diethylsulfamoyl)phenyl]carbonyl}amino)benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
3IL6
 
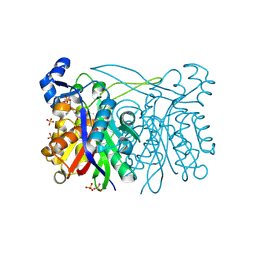 | | Structure of E. faecalis FabH in complex with 2-({4-[(3R,5S)-3,5-dimethylpiperidin-1-yl]-3-phenoxybenzoyl}amino)benzoic acid | | Descriptor: | 2-[({4-[(3R,5S)-3,5-dimethylpiperidin-1-yl]-3-phenoxyphenyl}carbonyl)amino]benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 3, SULFATE ION | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
3IL9
 
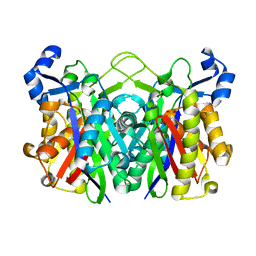 | | Structure of E. coli FabH | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
3IL7
 
 | | Crystal structure of S. aureus FabH | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
3IL3
 
 | | Structure of Haemophilus influenzae FabH | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
3IL4
 
 | | Structure of E. faecalis FabH in complex with acetyl CoA | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, ACETYL COENZYME *A | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
8OR8
 
 | |
7QB3
 
 | |
5MDI
 
 | | Crystal structure of TDP-43 N-terminal domain at 2.1 A resolution | | Descriptor: | ACETATE ION, TAR DNA-binding protein 43 | | Authors: | Afroz, T, Hock, E.-M, Ernst, P, Foglieni, C, Jambeau, M, Gilhespy, L, Laferriere, F, Maniecka, Z, Plueckthun, A, Mittl, P, Paganetti, P, Allain, F.H.T, Polymenidou, M. | | Deposit date: | 2016-11-11 | | Release date: | 2017-07-05 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation.
Nat Commun, 8, 2017
|
|
6GE1
 
 | |
1FEQ
 
 | | NMR SOLUTION STRUCTURE OF THE ANTICODON OF TRNA(LYS3) WITH T6A MODIFICATION AT POSITION 37 | | Descriptor: | 5'-R(*GP*CP*AP*GP*AP*CP*UP*UP*UP*UP*(T6A)P*AP*UP*CP*UP*GP*C)-3' | | Authors: | Stuart, J.W, Gdaniec, Z, Guenther, R.H, Marszalek, M, Sochacka, E, Malkiewicz, A, Agris, P.F. | | Deposit date: | 2000-07-21 | | Release date: | 2000-11-22 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A.
Biochemistry, 39, 2000
|
|
1XV6
 
 | |
2JXS
 
 | |
2K41
 
 | | NMR structure of uridine bulged RNA duplex | | Descriptor: | RNA_(5'-R(*CP*AP*GP*CP*CP*GP*AP*C)-3')_, RNA_(5'-R(*GP*UP*CP*GP*UP*GP*CP*UP*G)-3')_ | | Authors: | Popenda, L, Bielecki, L, Gdaniec, Z, Adamiak, R.W. | | Deposit date: | 2008-05-26 | | Release date: | 2008-11-04 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of adenosine bulged RNA duplex reveals formation of the dinucleotide platform in the C:G-A triple
ARKIVOC, 2009, 2008
|
|
2K3Z
 
 | | NMR structure of adenosine bulged RNA duplex with C:G-A triple | | Descriptor: | RNA_(5'-R(*CP*AP*GP*CP*CP*GP*AP*C)-3')_, RNA_(5'-R(*GP*UP*CP*GP*AP*GP*CP*UP*G)-3')_ | | Authors: | Popenda, L, Bielecki, L, Gdaniec, Z, Adamiak, R.W. | | Deposit date: | 2008-05-26 | | Release date: | 2008-11-04 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of adenosine bulged RNA duplex reveals formation of the dinucleotide platform in the C:G-A triple
ARKIVOC, 2009, 2008
|
|
2JXQ
 
 | | NMR structure of RNA duplex | | Descriptor: | RNA (5'-R(*CP*GP*CP*UP*CP*UP*CP*UP*GP*C)-3'), RNA (5'-R(*GP*CP*AP*GP*AP*GP*AP*GP*CP*G)-3') | | Authors: | Popenda, L, Adamiak, R.W, Gdaniec, Z. | | Deposit date: | 2007-11-28 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Bulged adenosine influence on the RNA duplex conformation in solution.
Biochemistry, 47, 2008
|
|
