8EJZ
 
 | | [4+2] Aza-Cyclase Y293F variant | | Descriptor: | PbtD | | Authors: | Nair, S.K. | | Deposit date: | 2022-09-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enzymatic Pyridine Aromatization during Thiopeptide Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
4OK4
 
 | | Crystal Structure of Alg17c Mutant H202L | | Descriptor: | Putative alginate lyase, ZINC ION | | Authors: | Nair, S.K, Park, D.S. | | Deposit date: | 2014-01-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a PL17 Family Alginate Lyase Demonstrates Functional Similarities among Exotype Depolymerases.
J.Biol.Chem., 289, 2014
|
|
4OJZ
 
 | |
4OK2
 
 | | Crystal Structure of Alg17c Mutant Y258A | | Descriptor: | Putative alginate lyase, ZINC ION | | Authors: | Nair, S.K, Park, D.S. | | Deposit date: | 2014-01-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of a PL17 Family Alginate Lyase Demonstrates Functional Similarities among Exotype Depolymerases.
J.Biol.Chem., 289, 2014
|
|
3R9E
 
 | |
3R95
 
 | |
3R9F
 
 | |
3R9G
 
 | |
3R96
 
 | |
4H6W
 
 | |
4I3V
 
 | |
4H6V
 
 | |
4I3X
 
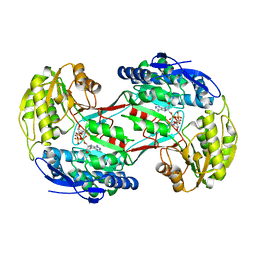 | |
4H6X
 
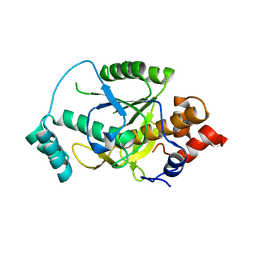 | | Structure of Patellamide maturation protease PatG | | Descriptor: | Thiazoline oxidase/subtilisin-like protease | | Authors: | Nair, S.K, Agarwal, V. | | Deposit date: | 2012-09-19 | | Release date: | 2012-10-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of cyanobactin maturation enzymes define a family of transamidating proteases.
Chem.Biol., 19, 2012
|
|
4I3U
 
 | |
4I3W
 
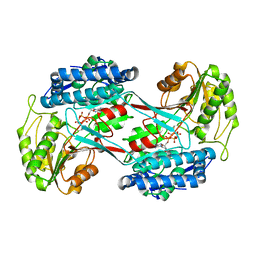 | |
4I3T
 
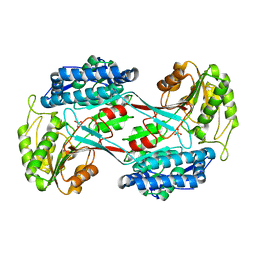 | |
2G02
 
 | | Nisin cyclase | | Descriptor: | Nisin biosynthesis protein nisC, ZINC ION | | Authors: | Nair, S.K. | | Deposit date: | 2006-02-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mechanism of the Lantibiotic Cyclase Involved in Nisin Biosynthesis
Science, 311, 2006
|
|
2G0D
 
 | | Nisin cyclase | | Descriptor: | Nisin biosynthesis protein nisC, ZINC ION | | Authors: | Nair, S.K. | | Deposit date: | 2006-02-12 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure and mechanism of the lantibiotic cyclase involved in nisin biosynthesis
Science, 311, 2006
|
|
3G7D
 
 | | Native PhpD with Cadmium Atoms | | Descriptor: | CADMIUM ION, PhpD | | Authors: | Nair, S.K. | | Deposit date: | 2009-02-09 | | Release date: | 2009-06-09 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An unusual carbon-carbon bond cleavage reaction during phosphinothricin biosynthesis.
Nature, 459, 2009
|
|
5UW5
 
 | | PCY1 H695A Variant in Complex with Follower Peptide | | Descriptor: | CACODYLATE ION, CALCIUM ION, Peptide cyclase 1, ... | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Characterization of the macrocyclase involved in the biosynthesis of RiPP cyclic peptides in plants.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UW7
 
 | | PCY1 Y481F Variant in Complex with Follower Peptide | | Descriptor: | MAGNESIUM ION, Peptide cyclase 1, Presegetalin A1 | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Characterization of the macrocyclase involved in the biosynthesis of RiPP cyclic peptides in plants.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4NEI
 
 | | Alg17c PL17 Family Alginate Lyase | | Descriptor: | ZINC ION, alginate lyase | | Authors: | Park, D.S, Nair, S.K. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a PL17 Family Alginate Lyase Demonstrates Functional Similarities among Exotype Depolymerases.
J.Biol.Chem., 289, 2014
|
|
7QOS
 
 | |
7RTY
 
 | |
