7Y3A
 
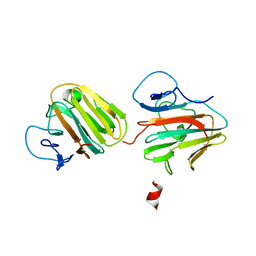 | | Crystal structure of TRIM7 bound to 2C | | Descriptor: | E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,TRIM7-2C | | Authors: | Dong, C, Yan, X. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XYU
 
 | |
7XYX
 
 | |
7XYW
 
 | |
7XYV
 
 | |
7XYS
 
 | |
7YNX
 
 | | Crystal structure of Pirh2 bound to poly-Ala peptide | | Descriptor: | RING finger and CHY zinc finger domain-containing protein 1, SODIUM ION, SULFATE ION, ... | | Authors: | Dong, C, Yan, X, Li, Y. | | Deposit date: | 2022-08-01 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of an Ala-rich C-degron by the E3 ligase Pirh2.
Nat Commun, 14, 2023
|
|
8J4A
 
 | | Crystal structure of OY phytoplasma SAP05 in complex with AtRPN10 | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Nat Commun, 15, 2024
|
|
8J49
 
 | | Crystal structure of OY phytoplasma SAP05 in complex with AtSPL5 | | Descriptor: | Sequence-variable mosaic (SVM) signal sequence domain-containing protein, Squamosa promoter-binding-like protein 5, ZINC ION | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Nat Commun, 15, 2024
|
|
8J4B
 
 | | Crystal structure of OY phytoplasma SAP05 in complex with AtSPL13 | | Descriptor: | Sequence-variable mosaic (SVM) signal sequence domain-containing protein, Squamosa promoter-binding-like protein 13A, ZINC ION | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Nat Commun, 15, 2024
|
|
8J48
 
 | | Crystal structure of OY phytoplasma SAP05 in complex with AtGATA18 | | Descriptor: | GATA transcription factor 18, Sequence-variable mosaic (SVM) signal sequence domain-containing protein, ZINC ION | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Nat Commun, 15, 2024
|
|
7EP2
 
 | | Crystal structure of ZYG11B bound to GGFN degron | | Descriptor: | Protein zyg-11 homolog B | | Authors: | Yan, X, Li, Y. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Mol.Cell, 81, 2021
|
|
7EP5
 
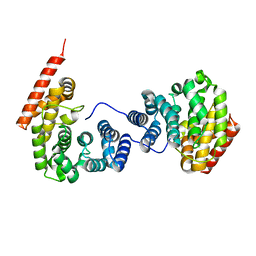 | | Crystal structure of ZER1 bound to GKLH degron | | Descriptor: | Protein zer-1 homolog | | Authors: | Yan, X, Li, Y. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Mol.Cell, 81, 2021
|
|
7EP0
 
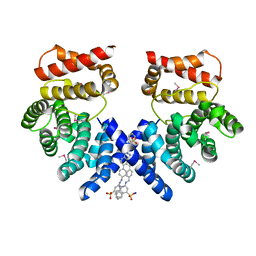 | | Crystal structure of ZYG11B bound to GSTE degron | | Descriptor: | Protein zyg-11 homolog B, sodium 3,3'-(1E,1'E)-biphenyl-4,4'-diylbis(diazene-2,1-diyl)bis(4-aminonaphthalene-1-sulfonate) | | Authors: | Yan, X, Li, Y. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-14 | | Last modified: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Mol.Cell, 81, 2021
|
|
7EP3
 
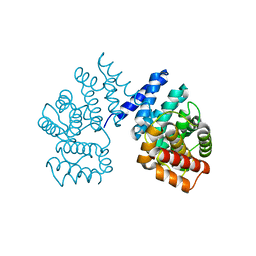 | | Crystal structure of ZER1 bound to GAGN degron | | Descriptor: | Protein zer-1 homolog | | Authors: | Yan, X, Li, Y. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.513 Å) | | Cite: | Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Mol.Cell, 81, 2021
|
|
7EP1
 
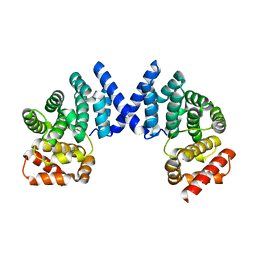 | | Crystal structure of ZYG11B bound to GFLH degron | | Descriptor: | Protein zyg-11 homolog B | | Authors: | Yan, X, Li, Y. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Mol.Cell, 81, 2021
|
|
7EP4
 
 | | Crystal structure of ZER1 bound to GFLH degron | | Descriptor: | Protein zer-1 homolog | | Authors: | Yan, X, Li, Y. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Mol.Cell, 81, 2021
|
|
7JYA
 
 | | Crystal structure of E3 ligase in complex with peptide | | Descriptor: | ASN-ARG-ARG-ARG-ARG-TRP-ARG-GLU-ARG-GLN-ARG, Protein fem-1 homolog C, UNKNOWN ATOM OR ION | | Authors: | Yan, X, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Dong, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-30 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Molecular basis for ubiquitin ligase CRL2 FEM1C -mediated recognition of C-degron.
Nat.Chem.Biol., 17, 2021
|
|
