7UGH
 
 | |
4KMS
 
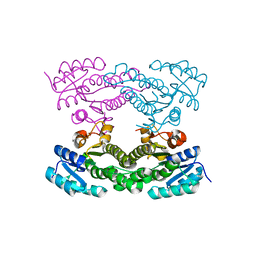 | | Crystal structure of Acetoacetyl-CoA reductase from Rickettsia felis | | Descriptor: | Acetoacetyl-CoA reductase | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Abendroth, J, Lukacs, C, Edwards, T.E, Lorimer, D. | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of acetoacetyl-CoA reductase from Rickettsia felis.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
8CU5
 
 | |
8CTR
 
 | |
8CU9
 
 | |
8CSO
 
 | |
8CXA
 
 | |
8DIM
 
 | |
8DIU
 
 | |
8DIS
 
 | |
8DU0
 
 | |
8DTE
 
 | |
8DTC
 
 | |
8DU1
 
 | |
8DT6
 
 | |
8DUY
 
 | |
8DV0
 
 | |
8F38
 
 | |
3PGX
 
 | |
3PXX
 
 | |
3S55
 
 | |
3SX2
 
 | |
3T7C
 
 | |
3TK1
 
 | |
3TSC
 
 | |
