3MD0
 
 | |
5EJ2
 
 | |
3NXS
 
 | |
3OEC
 
 | |
7SBC
 
 | |
7SBJ
 
 | |
7SIQ
 
 | |
7SIR
 
 | |
7SKB
 
 | |
7SZP
 
 | |
7SY9
 
 | |
7T35
 
 | |
7T7J
 
 | |
7TCM
 
 | |
7TI7
 
 | |
7TJ3
 
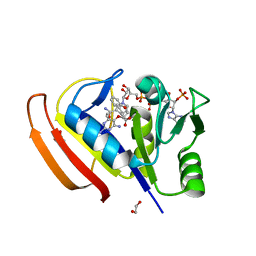 | |
7SSR
 
 | | Crystal Structure of Ebola zaire Envelope glycoprotein GP in complex with compound ARN0075093 | | Descriptor: | (1R,2s,3S,5s,7s)-N-[(1r,4r)-4-(aminomethyl)cyclohexyl]-5-phenyladamantane-2-carboxamide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-11-11 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Ebola zaire Envelope glycoprotein GP in complex with compound ARN0075093
to be published
|
|
7SSQ
 
 | |
5JRY
 
 | |
5K9F
 
 | |
5JSC
 
 | |
5KF0
 
 | |
5KIA
 
 | |
5KHA
 
 | |
5KOI
 
 | |
