4XHV
 
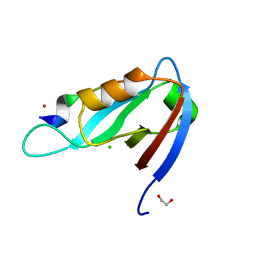 | | Crystal structure of Drosophila Spinophilin-PDZ and a C-terminal peptide of Neurexin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, LP20995p, ... | | Authors: | Driller, J.H, Muhammad, K.G.H, Reddy, S, Rey, U, Boehme, M.A, Hollmann, C, Ramesh, N, Depner, H, Luetzkendorf, J, Matkovic, T, Bergeron, D, Quentin, C, Schmoranzer, J, Goettfert, F, Holt, M, Wahl, M.C, Hell, S.W, Walter, A, Sigrist, S.J, Loll, B. | | Deposit date: | 2015-01-06 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Presynaptic spinophilin tunes neurexin signalling to control active zone architecture and function.
Nat Commun, 6, 2015
|
|
1W5C
 
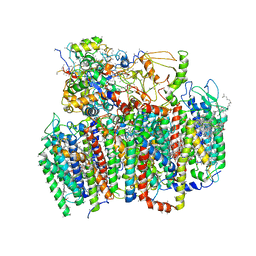 | | Photosystem II from Thermosynechococcus elongatus | | Descriptor: | 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Biesiadka, J, Loll, B, Kern, J, Irrgang, K.-D, Saenger, W. | | Deposit date: | 2004-08-06 | | Release date: | 2004-12-21 | | Last modified: | 2014-01-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of Cyanobacterial Photosystem II at 3.2 A Resolution: A Closer Look at the Mn- Cluster
Phys.Chem.Chem.Phys., 6, 2004
|
|
7R3W
 
 | |
6H5T
 
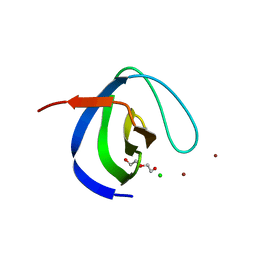 | | Intersectin SH3A short isoform | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Driller, J.H, Gerth, F, Freund, C, Wahl, M.C, Loll, B. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | Exon Inclusion Modulates Conformational Plasticity and Autoinhibition of the Intersectin 1 SH3A Domain.
Structure, 27, 2019
|
|
5G27
 
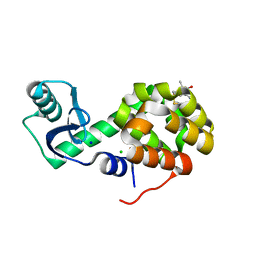 | | Structure of Spin-labelled T4 lysozyme mutant L118C-R1 at Room Temperature | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, ENDOLYSIN, ... | | Authors: | Gohlke, U, Consentius, P, Loll, B, Mueller, R, Kaupp, M, Heinemann, U, Risse, T. | | Deposit date: | 2016-04-07 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Tracking Transient Conformational States of T4 Lysozyme at Room Temperature Combining X-Ray Crystallography and Site-Directed Spin Labeling.
J.Am.Chem.Soc., 138, 2016
|
|
4Z8A
 
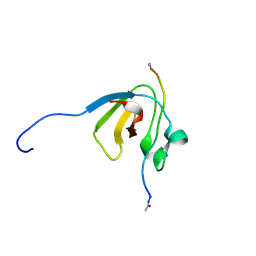 | | SH3-III of Drosophila Rim-binding protein bound to a Cacophony derived peptide | | Descriptor: | RIM-binding protein, isoform F, Voltage-dependent calcium channel type A subunit alpha-1 | | Authors: | Driller, J.H, Holton, N, Siebert, M, Boehme, A.M, Wahl, M.C, Sigrist, S.J, Loll, B. | | Deposit date: | 2015-04-08 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | A high affinity RIM-binding protein/Aplip1 interaction prevents the formation of ectopic axonal active zones.
Elife, 4, 2015
|
|
4Z88
 
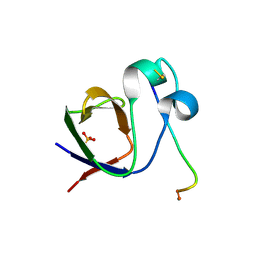 | | SH3-II of Drosophila Rim-binding protein with Aplip1 peptide | | Descriptor: | JNK-interacting protein 1, PHOSPHATE ION, RIM-binding protein, ... | | Authors: | Driller, J.H, Holton, N, Siebert, M, Boehme, M.A, Wahl, M.C, Sigrist, S.J, Loll, B. | | Deposit date: | 2015-04-08 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A high affinity RIM-binding protein/Aplip1 interaction prevents the formation of ectopic axonal active zones.
Elife, 4, 2015
|
|
7PLQ
 
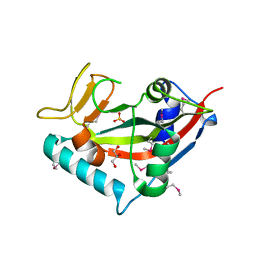 | | Crystal structure of the PARP domain of wheat SRO1 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, ... | | Authors: | Wirthmueller, L, Loll, B. | | Deposit date: | 2021-09-01 | | Release date: | 2021-10-27 | | Last modified: | 2022-11-09 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The superior salinity tolerance of bread wheat cultivar Shanrong No. 3 is unlikely to be caused by elevated Ta-sro1 poly-(ADP-ribose) polymerase activity.
Plant Cell, 34, 2022
|
|
4Z89
 
 | | SH3-II of Drosophila Rim-binding protein bound to a Cacophony derived peptide | | Descriptor: | CALCIUM ION, RIM-binding protein, isoform F, ... | | Authors: | Driller, J.H, Holton, N, Siebert, M, Boehme, M.A, Wahl, M.C, Sigrist, S.J, Loll, B. | | Deposit date: | 2015-04-08 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | A high affinity RIM-binding protein/Aplip1 interaction prevents the formation of ectopic axonal active zones.
Elife, 4, 2015
|
|
6TM8
 
 | | Crystal structure of glycoprotein D of Equine Herpesvirus Type 4 | | Descriptor: | Envelope glycoprotein D, GLYCEROL | | Authors: | Kremling, V, Loll, B, Osterrieder, N, Wahl, M, Dahmani, I, Chiantia, P, Azab, W. | | Deposit date: | 2019-12-03 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of glycoprotein D of equine alphaherpesviruses reveal potential binding sites to the entry receptor MHC-I.
Front Microbiol, 14, 2023
|
|
6TEO
 
 | | Crystal structure of a yeast Snu114-Prp8 complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Pre-mRNA-splicing factor 8, ... | | Authors: | Ganichkin, O, Jia, J, Loll, B, Absmeier, E, Wahl, M.C. | | Deposit date: | 2019-11-12 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Snu114-GTP-Prp8 module forms a relay station for efficient splicing in yeast.
Nucleic Acids Res., 48, 2020
|
|
6TQN
 
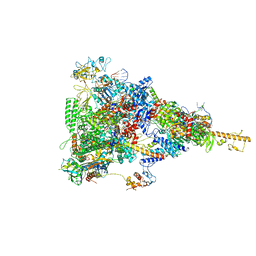 | | rrn anti-termination complex without S4 | | Descriptor: | 30S ribosomal protein S10, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Huang, Y.H, Wahl, M.C, Loll, B, Hilal, T, Said, N. | | Deposit date: | 2019-12-17 | | Release date: | 2020-08-05 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-Based Mechanisms of a Molecular RNA Polymerase/Chaperone Machine Required for Ribosome Biosynthesis.
Mol.Cell, 79, 2020
|
|
6TQO
 
 | | rrn anti-termination complex | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S4, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Huang, Y.H, Wahl, M.C, Loll, B, Hilal, T, Said, N. | | Deposit date: | 2019-12-17 | | Release date: | 2020-08-05 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-Based Mechanisms of a Molecular RNA Polymerase/Chaperone Machine Required for Ribosome Biosynthesis.
Mol.Cell, 79, 2020
|
|
5ODO
 
 | | Crystal Structure of the Oleate hydratase of Rhodococcus erythropolis | | Descriptor: | FORMIC ACID, GLYCEROL, Isomerase, ... | | Authors: | Driller, R, Lorenzen, J, Waldow, A, Qoura, F, Brueck, T, Loll, B. | | Deposit date: | 2017-07-06 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Rhodococcus erythropolis Oleate Hydratase: a New Member in the Oleate Hydratase Family Tree - Biochemical and Structural Studies.
Chemcatchem, 2017
|
|
5NWS
 
 | | Crystal structure of saAcmM involved in actinomycin biosynthesis | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, TETRAETHYLENE GLYCOL, ... | | Authors: | Driller, R, Semsary, S, Crnovicic, I, Vater, J, Keller, U, Loll, B. | | Deposit date: | 2017-05-08 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Ketonization of Proline Residues in the Peptide Chains of Actinomycins by a 4-Oxoproline Synthase.
Chembiochem, 19, 2018
|
|
5NX0
 
 | | Structure of Spin-labelled T4 lysozyme mutant L115C-R119C-R1 at room temperature | | Descriptor: | Endolysin | | Authors: | Gohlke, U, Loll, B, Consentius, P, Mueller, R, Kaupp, M, Heinemann, U, Wahl, M.C, Risse, T. | | Deposit date: | 2017-05-09 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Combining EPR spectroscopy and X-ray crystallography to elucidate the structure and dynamics of conformationally constrained spin labels in T4 lysozyme single crystals.
Phys Chem Chem Phys, 19, 2017
|
|
4NAO
 
 | | Crystal structure of EasH | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Janke, R, Havemann, J, Vogel, D, Keller, U, Loll, B. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Cyclolization of d-lysergic Acid alkaloid peptides.
Chem.Biol., 21, 2014
|
|
6IB8
 
 | | Structure of a complex of SuhB and NusA AR2 domain | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, Inositol-1-monophosphatase, ... | | Authors: | Huang, Y.H, Loll, B, Wahl, M.C. | | Deposit date: | 2018-11-29 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Structural basis for the function of SuhB as a transcription factor in ribosomal RNA synthesis.
Nucleic Acids Res., 47, 2019
|
|
6GGJ
 
 | | Crystal structure of CotB2 in complex with alendronate | | Descriptor: | 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE, Cyclooctat-9-en-7-ol synthase, MAGNESIUM ION | | Authors: | Driller, R, Janke, S, Fuchs, M, Warner, E, Mhashal, A.R, Major, D.T, Christmann, M, Brueck, T, Loll, B. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards a comprehensive understanding of the structural dynamics of a bacterial diterpene synthase during catalysis.
Nat Commun, 9, 2018
|
|
6IB7
 
 | | Structure of wild type SuhB | | Descriptor: | GLYCEROL, Inositol-1-monophosphatase, MAGNESIUM ION | | Authors: | Huang, Y.H, Loll, B, Wahl, M.C. | | Deposit date: | 2018-11-29 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Structural basis for the function of SuhB as a transcription factor in ribosomal RNA synthesis.
Nucleic Acids Res., 47, 2019
|
|
6G70
 
 | | Structure of murine Prpf39 | | Descriptor: | Pre-mRNA-processing factor 39 | | Authors: | De Bortoli, F.D, Loll, B, Wahl, M, Heyd, F. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-03 | | Last modified: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Increased versatility despite reduced molecular complexity: evolution, structure and function of metazoan splicing factor PRPF39.
Nucleic Acids Res., 47, 2019
|
|
8QWR
 
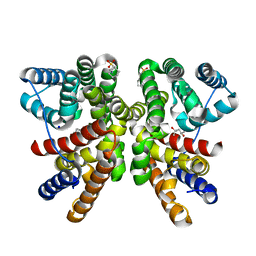 | | Crystal structure of CotB2 variant V80L | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Cyclooctat-9-en-7-ol synthase, ... | | Authors: | Dimos, N, Himpich, S, Ringel, M, Driller, R, Major, D.T, Brueck, T, Loll, B. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of CotB2 variant V80L
To Be Published
|
|
8QWS
 
 | | Crystal structure of CotB2 variant V80L in complex with alendronate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Dimos, N, Himpich, S, Ringel, M, Driller, R, Major, D.T, Brueck, T, Loll, B. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of CotB2 variant V80L in complex with alendronate
To Be Published
|
|
6Z9T
 
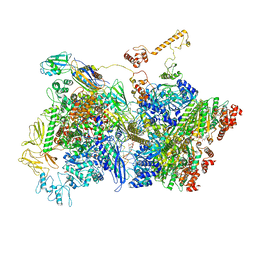 | | Transcription termination intermediate complex 5 | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-04 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
6Z9P
 
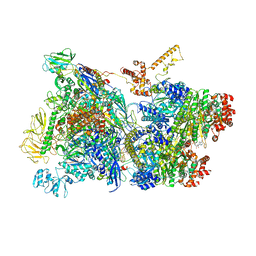 | | Transcription termination intermediate complex 1 | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-04 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
