4C40
 
 | |
5FK1
 
 | |
6YL5
 
 | | Crystal structure of the SAM-SAH riboswitch with SAH | | Descriptor: | Chains: A,B,C,D,E,F,G,H,I,J,K,L, MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 2020
|
|
6YMI
 
 | | Crystal structure of the SAM-SAH riboswitch with AMP. | | Descriptor: | 5-BROMOCYTIDINE 5'-(DIHYDROGEN PHOSPHATE), ADENOSINE MONOPHOSPHATE, Chains: A,C,F,I,M,O, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 48, 2020
|
|
6YMJ
 
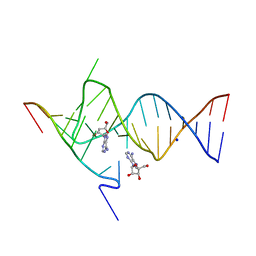 | | Crystal structure of the SAM-SAH riboswitch with adenosine. | | Descriptor: | 5-BROMOCYTIDINE 5'-(DIHYDROGEN PHOSPHATE), ADENOSINE, Chains: A,C,F,I,M,O, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 48, 2020
|
|
6YLB
 
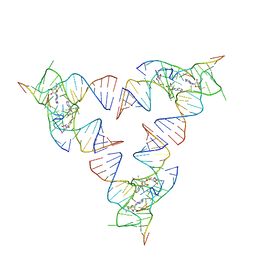 | | Crystal structure of the SAM-SAH riboswitch with SAM | | Descriptor: | Chains: A,C,F,I,M,O, Chains: B,D,G,J,N,P, S-ADENOSYLMETHIONINE | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 2020
|
|
6YML
 
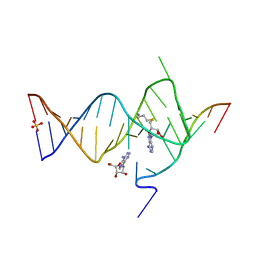 | |
6YMK
 
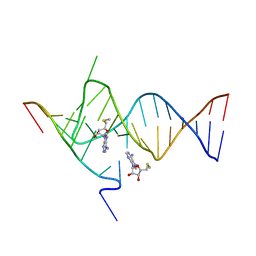 | | Crystal structure of the SAM-SAH riboswitch with AMP | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Chains: A,C,F,I,M,O, Chains: B,D,G,J,N,P, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 48, 2020
|
|
5NEF
 
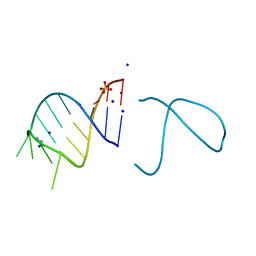 | | The structure of the G. violaceus guanidine II riboswitch P1 stem-loop with guanidine | | Descriptor: | GUANIDINE, RNA (5'-R(*GP*GP*UP*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*AP*(CBV)P*C)-3'), SODIUM ION, ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-03-10 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NDH
 
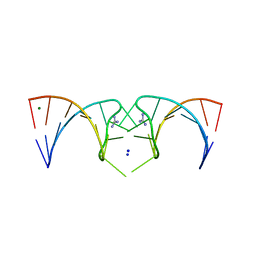 | | The structure of the G. violaceus guanidine II riboswitch P2 stem-loop | | Descriptor: | GUANIDINE, MAGNESIUM ION, RNA (5'-R(*GP*(CBV)P*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*GP*C)-3'), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
6FZ0
 
 | |
6HBX
 
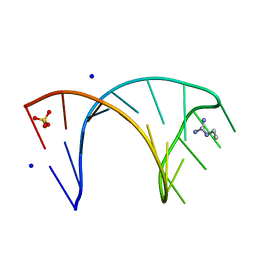 | |
6HBT
 
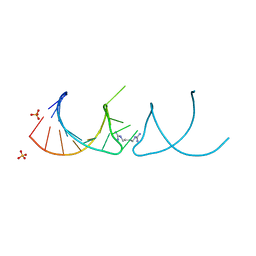 | |
6HCT
 
 | |
6HC5
 
 | |
6TFG
 
 | |
6TF1
 
 | |
6TFF
 
 | |
6TF0
 
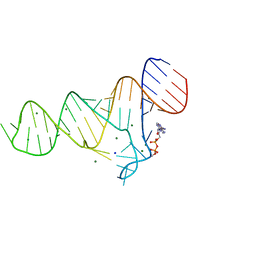 | | Crystal structure of the ADP-binding domain of the NAD+ riboswitch with Nicotinamide adenine dinucleotide, reduced (NADH) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Chains: A, MAGNESIUM ION, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2019-11-12 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and ligand binding of the ADP-binding domain of the NAD+ riboswitch.
Rna, 26, 2020
|
|
6TF3
 
 | |
6TFE
 
 | |
6TB7
 
 | |
6TF2
 
 | |
6TFH
 
 | |
5FK4
 
 | |
