8CKE
 
 | |
8CH3
 
 | | PBP AccA from A. vitis S4 in complex with Agrocinopine C-like | | Descriptor: | 2-O-phosphono-alpha-D-glucopyranose, Agrocinopine utilization periplasmic binding protein AccA, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CH2
 
 | | PBP AccA from A. vitis S4 in complex with L-arabinose-2-phosphate (A2P) | | Descriptor: | 1,2-ETHANEDIOL, 2-O-phosphono-alpha-L-arabinopyranose, 2-O-phosphono-beta-L-arabinopyranose, ... | | Authors: | Morera, S, Deicsics, G, Vigouroux, A. | | Deposit date: | 2023-02-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CDO
 
 | |
8C6R
 
 | | PBP AccA from A. tumefaciens Bo542 in apoform 4 | | Descriptor: | 1,2-ETHANEDIOL, Agrocinopine utilization periplasmic binding protein AccA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.884 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CJU
 
 | | PBP AccA from A. vitis S4 in complex with agrocin84 | | Descriptor: | ABC transporter substrate binding protein (Agrocinopine), [(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl]oxy-N-[9-[(2R,3S,5R)-5-[[[(2R,3S)-4-methyl-2,3-bis(oxidanyl)pentanoyl]amino]-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxolan-2-yl]purin-6-yl]phosphonamidic acid | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CKD
 
 | | PBP AccA from A. fabrum C58 in complex with agrocinopine D-like | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter substrate-binding protein, Agrocinopine D-like (C2-C2 linked; with an alpha and beta-D-glucopyranose), ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-15 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
5M2W
 
 | | Structure of nanobody nb18 raised against TssK from E. coli T6SS | | Descriptor: | Llama nanobody nb8 against TssK from T6SS, SULFATE ION | | Authors: | Cambillau, C, Nguyen, V.S, Spinelli, S, Desmyter, A. | | Deposit date: | 2016-10-13 | | Release date: | 2017-06-28 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex.
Nat Microbiol, 2, 2017
|
|
6SKJ
 
 | | DeltaC2 C-terminal truncation of HsNMT1 in complex with MyrCoA and GNCFSKPR substrates | | Descriptor: | Apoptosis-inducing factor 3, COENZYME A, GLYCEROL, ... | | Authors: | Dian, C, Riviere, F.B, Asensio, T, Giglione, C, Meinnel, T. | | Deposit date: | 2019-08-15 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
6SK8
 
 | | DeltaC3 C-terminal truncation of HsNMT1 in complex with MyrCoA and GDCFSKPR substrates | | Descriptor: | Apoptosis-inducing factor 3, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dian, C, Riviere, F.B, Asensio, T, Giglione, C, Meinnel, T. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
6SJZ
 
 | | HsNMT1 in complex with both MyrCoA and Acetylated-GNCFSKPR substrates | | Descriptor: | Apoptosis-inducing factor 3, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dian, C, Riviere, F.B, Asensio, T, Giglione, C, Meinnel, T. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
6SK3
 
 | | C-terminal HsNMT1 deltaC3 truncation in complex with both MyrCoA and GNCFSKPR substrates | | Descriptor: | Apoptosis-inducing factor 3, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Dian, C, Riviere, F.B, Asensio, T, Giglione, C, Meinnel, T. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
6TG3
 
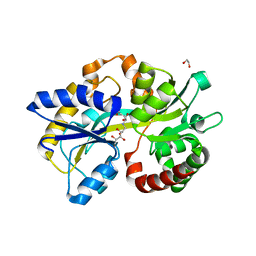 | | Crystal Structure of the PBP/SBP MotA in complex with glucopinic acid from A. tumefaciens B6/R10 | | Descriptor: | (2~{S})-2-[[(3~{S},4~{R},5~{R})-3,4,5,6-tetrakis(oxidanyl)-2-oxidanylidene-hexyl]amino]pentanedioic acid, 1,2-ETHANEDIOL, MotA | | Authors: | Morera, S, Vigouroux, S. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Import pathways of the mannityl-opines into the bacterial pathogen Agrobacterium tumefaciens: structural, affinity and in vivo approaches.
Biochem.J., 477, 2020
|
|
6TG2
 
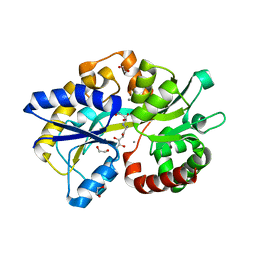 | | Structure of the PBP/SBP MotA in complex with mannopinic acid from A.tumefacien R10 | | Descriptor: | (2~{R})-2-[[(3~{R},4~{R},5~{S})-3,4,5,6-tetrakis(oxidanyl)-2-oxidanylidene-hexyl]amino]pentanedioic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Morera, S, Marty, L. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Import pathways of the mannityl-opines into the bacterial pathogen Agrobacterium tumefaciens: structural, affinity and in vivo approaches.
Biochem.J., 477, 2020
|
|
6TFS
 
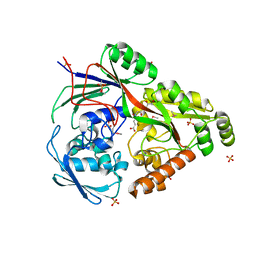 | | Structure in P3212 form of the PBP/SBP MoaA in complex with glucopinic acid from A.tumefacien R10 | | Descriptor: | (2~{S})-2-[[(3~{S},4~{R},5~{R})-3,4,5,6-tetrakis(oxidanyl)-2-oxidanylidene-hexyl]amino]pentanedioic acid, ABC transporter substrate-binding protein, CHLORIDE ION, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Import pathways of the mannityl-opines into the bacterial pathogen Agrobacterium tumefaciens: structural, affinity and in vivo approaches.
Biochem.J., 477, 2020
|
|
6TFQ
 
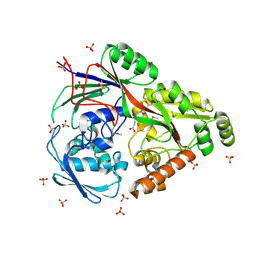 | | Structure in P3212 form of the PBP/SBP MoaA in complex with mannopinic acid from A.tumefacien R10 | | Descriptor: | (2~{R})-2-[[(3~{R},4~{R},5~{S})-3,4,5,6-tetrakis(oxidanyl)-2-oxidanylidene-hexyl]amino]pentanedioic acid, ABC transporter substrate-binding protein, CHLORIDE ION, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Import pathways of the mannityl-opines into the bacterial pathogen Agrobacterium tumefaciens: structural, affinity and in vivo approaches.
Biochem.J., 477, 2020
|
|
6TFX
 
 | | Structure in P21 form of the PBP/SBP MoaA in complex with mannopinic acid from A.tumefacien R10 | | Descriptor: | (2~{R})-2-[[(3~{R},4~{R},5~{S})-3,4,5,6-tetrakis(oxidanyl)-2-oxidanylidene-hexyl]amino]pentanedioic acid, 1,2-ETHANEDIOL, ABC transporter substrate-binding protein, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Import pathways of the mannityl-opines into the bacterial pathogen Agrobacterium tumefaciens: structural, affinity and in vivo approaches.
Biochem.J., 477, 2020
|
|
6EHJ
 
 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA and peptide bound | | Descriptor: | ASPARAGINE, COENZYME A, GLYCEROL, ... | | Authors: | Perez-Dorado, I, Ritzefeld, M, Tate, E.W. | | Deposit date: | 2017-09-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
4NLB
 
 | |
4NLC
 
 | |
7PU5
 
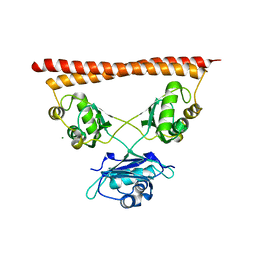 | | Structure of SFPQ-NONO complex | | Descriptor: | MAGNESIUM ION, Non-POU domain-containing octamer-binding protein, Splicing factor, ... | | Authors: | Fribourg, S. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Crystal structure of SFPQ-NONO heterodimer.
Biochimie, 198, 2022
|
|
7Q5A
 
 | | Lanreotide nanotube | | Descriptor: | Lanreotide | | Authors: | Pieri, L, Wang, F, Arteni, A.A, Bressanelli, S, Egelman, E.H, Paternostre, M. | | Deposit date: | 2021-11-03 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Atomic structure of Lanreotide nanotubes revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6H6O
 
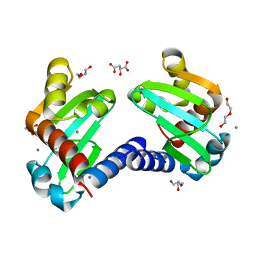 | | UbiJ-SCP2 Ubiquinone synthesis protein | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fyfe, C.D, Legrand, P, Pecqueur, L, Ciccone, L, Lombard, M, Fontecave, M. | | Deposit date: | 2018-07-28 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Soluble Metabolon Synthesizes the Isoprenoid Lipid Ubiquinone.
Cell Chem Biol, 26, 2019
|
|
6H6N
 
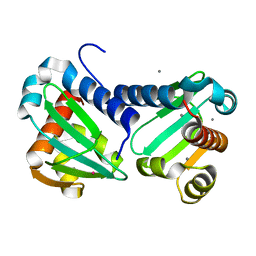 | | UbiJ-SCP2 Ubiquinone synthesis protein | | Descriptor: | CALCIUM ION, TERBIUM(III) ION, Ubiquinone biosynthesis protein UbiJ | | Authors: | Fyfe, C.D, Legrand, P, Pecqueur, L, Ciccone, L, Lombard, M, Fontecave, M. | | Deposit date: | 2018-07-28 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A Soluble Metabolon Synthesizes the Isoprenoid Lipid Ubiquinone.
Cell Chem Biol, 26, 2019
|
|
6H6P
 
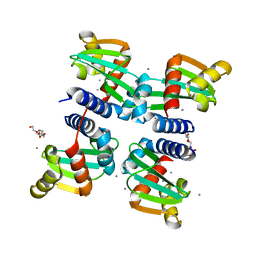 | | UbiJ-SCP2 Ubiquinone synthesis protein | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, Ubiquinone biosynthesis protein UbiJ | | Authors: | Fyfe, C.D, Legrand, P, Pecqueur, L, Ciccone, L, Lombard, M, Fontecave, M. | | Deposit date: | 2018-07-28 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Soluble Metabolon Synthesizes the Isoprenoid Lipid Ubiquinone.
Cell Chem Biol, 26, 2019
|
|
