3AEQ
 
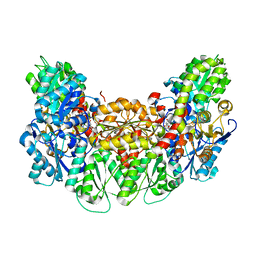 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N, ... | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
3AES
 
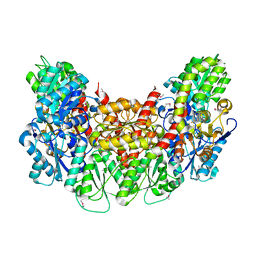 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
3AB1
 
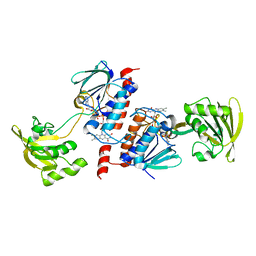 | | Crystal Structure of Ferredoxin NADP+ Oxidoreductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase | | Authors: | Muraki, N, Seo, D, Kurisu, G. | | Deposit date: | 2009-11-30 | | Release date: | 2010-11-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Asymmetric dimeric structure of ferredoxin-NAD(P)+ oxidoreductase from the green sulfur bacterium Chlorobaculum tepidum: implications for binding ferredoxin and NADP+
J.Mol.Biol., 401, 2010
|
|
3AJV
 
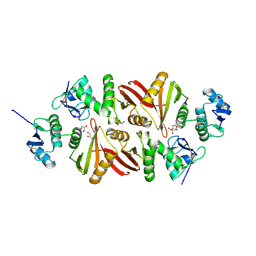 | | Splicing endonuclease from Aeropyrum pernix | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative uncharacterized protein, ... | | Authors: | Yoshinari, S, Watanabe, Y, Okuda, M, Shiba, T, Inaoka, K.D, Kurisu, G. | | Deposit date: | 2010-06-19 | | Release date: | 2010-11-17 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Conserved Lysine Residue in the Crenarchaea-Specific Loop is Important for the Crenarchaeal Splicing Endonuclease Activity.
J.Mol.Biol., 405, 2011
|
|
3AEK
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N, ... | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
3AET
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
3AEU
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
3AER
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
7XHX
 
 | | Crystal structure of metallo-beta-lactamase IMP-6 | | Descriptor: | Beta-lactamase, ZINC ION | | Authors: | Yamamoto, K, Tanaka, H, Kurisu, G, Nakano, R, Yano, H, Sakai, H. | | Deposit date: | 2022-04-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the substrate specificity of IMP-6 and IMP-1 metallo-beta-lactamases.
J.Biochem., 173, 2022
|
|
7XHW
 
 | | Crystal structure of metallo-beta-lactamase IMP-1 | | Descriptor: | Beta-lactamase, ZINC ION | | Authors: | Yamamoto, K, Tanaka, H, Kurisu, G, Nakano, R, Yano, H, Sakai, H. | | Deposit date: | 2022-04-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural insights into the substrate specificity of IMP-6 and IMP-1 metallo-beta-lactamases.
J.Biochem., 173, 2022
|
|
3VO2
 
 | |
3VKG
 
 | | X-ray structure of an MTBD truncation mutant of dynein motor domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dynein heavy chain, cytoplasmic, ... | | Authors: | Kon, T, Oyama, T, Shimo-Kon, R, Suto, K, Kurisu, G. | | Deposit date: | 2011-11-16 | | Release date: | 2012-03-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The 2.8 A crystal structure of the dynein motor domain
Nature, 484, 2012
|
|
3VO1
 
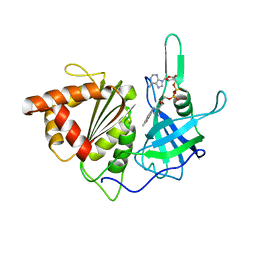 | |
3VV4
 
 | | Crystal structure of cyanobacteriochrome TePixJ GAF domain | | Descriptor: | Methyl-accepting chemotaxis protein, Phycoviolobilin, green light-absorbing form, ... | | Authors: | Ishizuka, T, Narikawa, R, Muraki, N, Shiba, T, Kurisu, G, Ikeuchi, M. | | Deposit date: | 2012-07-14 | | Release date: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of cyanobacteriochromes from phototaxis regulators AnPixJ and TePixJ reveal general and specific photoconversion mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3W2Z
 
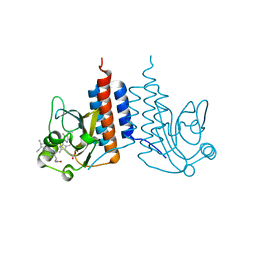 | | Crystal structure of the cyanobacterial protein | | Descriptor: | IODIDE ION, Methyl-accepting chemotaxis protein, PHYCOCYANOBILIN | | Authors: | Narikawa, R, Muraki, N, Shiba, T, Kurisu, G, Ikeuchi, M. | | Deposit date: | 2012-12-06 | | Release date: | 2013-01-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of cyanobacteriochromes from phototaxis regulators AnPixJ and TePixJ reveal general and specific photoconversion mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3W5U
 
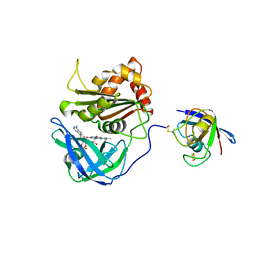 | | Cross-linked complex between Ferredoxin and Ferredoxin-NADP+ reductase | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin, ... | | Authors: | Kimata-Ariga, Y, Kubota-Kawai, H, Muraki, N, Hase, T, Kurisu, G. | | Deposit date: | 2013-02-06 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Concentration-dependent oligomerization of cross-linked complexes between ferredoxin and ferredoxin-NADP(+) reductase
Biochem.Biophys.Res.Commun., 434, 2013
|
|
3W6Q
 
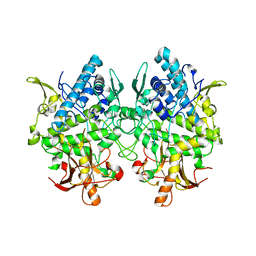 | | Crystal structure of melB apo-protyrosinase from Asperugillus oryzae | | Descriptor: | tyrosinase | | Authors: | Fujieda, N, Yabuta, S, Ikeda, T, Oyama, T, Muraki, N, Kurisu, G, Itoh, S. | | Deposit date: | 2013-02-20 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal structures of copper-depleted and copper-bound fungal pro-tyrosinase: insights into endogenous cysteine-dependent copper incorporation.
J.Biol.Chem., 288, 2013
|
|
3W5V
 
 | | Cross-linked complex between Ferredoxin and Ferredoxin-NADP+ reductase | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin, ... | | Authors: | Kimata-Ariga, Y, Kubota-Kawai, H, Muraki, N, Hase, T, Kurisu, G. | | Deposit date: | 2013-02-06 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | Concentration-dependent oligomerization of cross-linked complexes between ferredoxin and ferredoxin-NADP(+) reductase
Biochem.Biophys.Res.Commun., 434, 2013
|
|
3W6W
 
 | | Crystal structure of melB holo-protyrosinase from Asperugillus oryzae | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Fujieda, N, Yabuta, S, Ikeda, T, Oyama, T, Muraki, N, Kurisu, G, Itoh, S. | | Deposit date: | 2013-02-22 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.394 Å) | | Cite: | Crystal structures of copper-depleted and copper-bound fungal pro-tyrosinase: insights into endogenous cysteine-dependent copper incorporation.
J.Biol.Chem., 288, 2013
|
|
3WUQ
 
 | | Structure of the entire stalk region of the dynein motor domain | | Descriptor: | Cytoplasmic dynein 1 heavy chain 1 | | Authors: | Nishikawa, Y, Oyama, T, Kamiya, N, Kon, T, Toyoshima, Y.Y, Nakamura, H, Kurisu, G. | | Deposit date: | 2014-05-01 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the entire stalk region of the Dynein motor domain
J.Mol.Biol., 426, 2014
|
|
5BX3
 
 | | Crystal structure of Thermoanaerobacterium xylanolyticum GH116 beta-glucosidase with deoxynojirimycin | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYNOJIRIMYCIN, CALCIUM ION, ... | | Authors: | Charoenwattanasatien, R, Pengthaisong, S, Sansenya, S, Mutoh, R, Tankrathok, A, Tanaka, H, Kurisu, G, Ketudat Cairns, J.R. | | Deposit date: | 2015-06-08 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Bacterial beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (GBA2)
Acs Chem.Biol., 11, 2016
|
|
5BVU
 
 | | Crystal structure of Thermoanaerobacterium xylolyticum GH116 beta-glucosidase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Charoenwattanasatien, R, Pengthaisong, S, Sansenya, S, Mutoh, R, Tanaka, H, Kurisu, G, Ketudat Cairns, J.R. | | Deposit date: | 2015-06-05 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Bacterial beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (GBA2).
Acs Chem.Biol., 11, 2016
|
|
5BYR
 
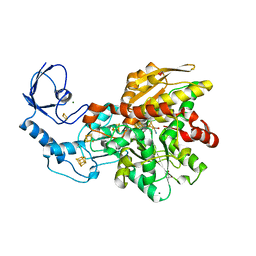 | | Semisynthetic [FeFe]-hydrogenase CpI with propane-dithiolato-bridged [2Fe] cofactor | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Esselborn, J, Muraki, N, Engelbrecht, V, Hofmann, E, Kurisu, G, Happe, T. | | Deposit date: | 2015-06-10 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A structural view of synthetic cofactor integration into [FeFe]-hydrogenases.
Chem Sci, 7, 2016
|
|
5BX5
 
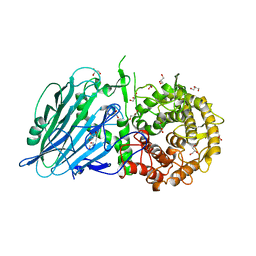 | | Crystal structure of Thermoanaerobacterium xylanolyticum GH116 beta-glucosidase with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Charoenwattanasatien, R, Pengthaisong, S, Sansenya, S, Mutoh, R, Hua, Y, Tanaka, H, Kurisu, G, Ketudat Cairns, J.R. | | Deposit date: | 2015-06-08 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bacterial beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (GBA2)
Acs Chem.Biol., 11, 2016
|
|
5BX4
 
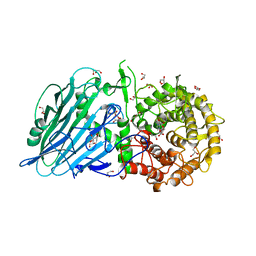 | | Crystal structure of Thermoanaerobacterium xylanolyticum GH116 beta-glucosidase with Glucoimidazole | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLUCOIMIDAZOLE, ... | | Authors: | Charoenwattanasatien, R, Pengthaisong, S, Sansenya, S, Mutoh, R, Tankrathok, A, Tanaka, H, Kurisu, G, Ketudat Cairns, J.R. | | Deposit date: | 2015-06-08 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Bacterial beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (GBA2)
Acs Chem.Biol., 11, 2016
|
|
