3ANG
 
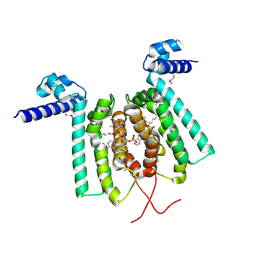 | | Crystal structure of Thermus thermophilus FadR in complex with E. coli-derived dodecyl-CoA | | Descriptor: | DODECYL-COA, Transcriptional repressor, TetR family | | Authors: | Agari, Y, Sakamoto, K, Agari, K, Kuramitsu, S, Shinkai, A. | | Deposit date: | 2010-09-01 | | Release date: | 2011-03-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | TetR-family transcriptional repressor Thermus thermophilus FadR controls fatty acid degradation.
Microbiology, 157, 2011
|
|
3AKC
 
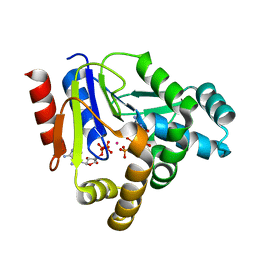 | | Crystal structure of CMP kinase in complex with CDP and ADP from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CYTIDINE-5'-DIPHOSPHATE, Cytidylate kinase, ... | | Authors: | Mega, R, Nakagawa, N, Kuramitsu, S, Masui, R. | | Deposit date: | 2010-07-12 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of the tertiary complex of CMP kinase with a phosphoryl group acceptor and a donor from Thermus thermophilus HB8
To be Published
|
|
3ANP
 
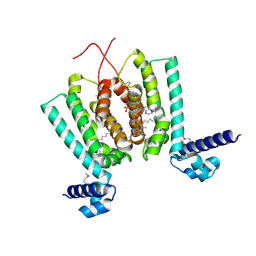 | | Crystal structure of Thermus thermophilus FadR, a TetR familly transcriptional repressor, in complex with lauroyl-CoA. | | Descriptor: | DODECYL-COA, LAURIC ACID, Transcriptional repressor, ... | | Authors: | Agari, Y, Agari, K, Sakamoto, K, Kuramitsu, S, Shinkai, A. | | Deposit date: | 2010-09-06 | | Release date: | 2011-03-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | TetR-family transcriptional repressor Thermus thermophilus FadR controls fatty acid degradation.
Microbiology, 157, 2011
|
|
3ASZ
 
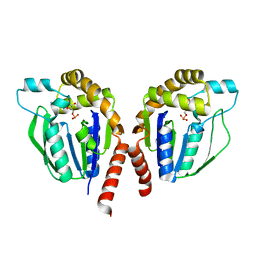 | | CMP-complex structure of uridine kinase from Thermus thermophilus HB8 | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Uridine kinase | | Authors: | Tomoike, F, Nakagawa, N, Kuramitsu, S, Masui, R. | | Deposit date: | 2010-12-22 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Single Amino Acid Limits the Substrate Specificity of Thermus thermophilus Uridine-Cytidine Kinase to Cytidine
Biochemistry, 50, 2011
|
|
3ASY
 
 | |
3B02
 
 | | Crystal structure of TTHB099, a transcriptional regulator CRP family from Thermus thermophilus HB8 | | Descriptor: | Transcriptional regulator, Crp family | | Authors: | Agari, Y, Kuramitsu, S, Shinkai, A, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-06-03 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | X-ray crystal structure of TTHB099, a CRP/FNR superfamily transcriptional regulator from Thermus thermophilus HB8, reveals a DNA-binding protein with no required allosteric effector molecule
Proteins, 80, 2012
|
|
2YWR
 
 | | Crystal structure of GAR transformylase from Aquifex aeolicus | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structures and reaction mechanisms of the two related enzymes, PurN and PurU.
J.Biochem., 154, 2013
|
|
2ZW2
 
 | | Crystal Structure of Formylglycinamide Ribonucleotide Amidotransferase III from SULFOLOBUS TOKODAII (STPURS) | | Descriptor: | GLYCEROL, Putative uncharacterized protein STS178 | | Authors: | Suzuki, S, Tamura, S, Okada, K, Baba, S, Kumasaka, T, Nakagawa, N, Masui, R, Kuramitsu, S, Sampei, G, Kawai, G. | | Deposit date: | 2008-11-27 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Formylglycinamide Ribonucleotide Amidotransferase III from SULFOLOBUS TOKODAII (STPURS)
To be Published
|
|
2ZDC
 
 | | Crystal structure of dUTPase from Sulfolobus tokodaii | | Descriptor: | 167aa long hypothetical dUTPase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION | | Authors: | Kanagawa, M, Baba S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-11-21 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of dUTPase from Sulfolobus tokodaii
To be Published
|
|
1UEB
 
 | | Crystal structure of translation elongation factor P from Thermus thermophilus HB8 | | Descriptor: | elongation factor P | | Authors: | Hanawa-Suetsugu, K, Sekine, S, Sakai, H, Hori-Takemoto, C, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-09 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of elongation factor P from Thermus thermophilus HB8
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1UEK
 
 | | Crystal structure of 4-(cytidine 5'-diphospho)-2C-methyl-D-erythritol kinase | | Descriptor: | 4-(cytidine 5'-diphospho)-2C-methyl-D-erythritol kinase | | Authors: | Wada, T, Kuramitsu, S, Yokoyama, S, Tame, J.R.H, Park, S.Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-17 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of 4-(Cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase, an Enzyme in the Non-mevalonate Pathway of Isoprenoid Synthesis.
J.Biol.Chem., 278, 2003
|
|
2YYU
 
 | | Crystal structure of uncharacterized conserved protein from Geobacillus kaustophilus | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Kanagawa, M, Baba, S, Nakamura, Y, Bessho, Y, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-02 | | Release date: | 2007-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of uncharacterized conserved protein from Geobacillus kaustophilus
To be Published
|
|
2Z02
 
 | | Crystal structure of phosphoribosylaminoimidazolesuccinocarboxamide synthase wit ATP from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, Phosphoribosylaminoimidazole-succinocarboxamide synthase, ... | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of phosphoribosylaminoimidazolesuccinocarboxamide synthase from Methanocaldococcus jannaschii
To be Published
|
|
2YWC
 
 | | Crystal structure of GMP synthetase from Thermus thermophilus in complex with XMP | | Descriptor: | GMP synthase [glutamine-hydrolyzing], XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-20 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of GMP synthetase from Thermus thermophilus
To be Published
|
|
2YWX
 
 | | Crystal structure of phosphoribosylaminoimidazole carboxylase catalytic subunit from Methanocaldococcus jannaschii | | Descriptor: | Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of phosphoribosylaminoimidazole carboxylase catalytic subunit from Methanocaldococcus jannaschii
To be Published
|
|
2YX5
 
 | | Crystal Structure of Methanocaldococcus jannaschii PurS, One of the Subunits of Formylglycinamide Ribonucleotide Amidotransferase in the Purine Biosynthetic Pathway | | Descriptor: | UPF0062 protein MJ1593 | | Authors: | Kanagawa, M, Baba, S, Agari, Y, Chen, L.Q, Fu, Z.-Q, Chrzas, J, Wang, B.C, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-24 | | Release date: | 2007-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Methanocaldococcus jannaschii PurS, One of the Subunits of Formylglycinamide Ribonucleotide Amidotransferase in the Purine Biosynthetic Pathway
To be Published
|
|
2YYH
 
 | |
2YZL
 
 | | Crystal structure of phosphoribosylaminoimidazole-succinocarboxamide synthase with ADP from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphoribosylaminoimidazole-succinocarboxamide synthase, SULFATE ION | | Authors: | Kanagawa, M, Baba, S, Fukui, K, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of phosphoribosylaminoimidazole-succinocarboxamide synthase with ADP from Methanocaldococcus jannaschii
To be Published
|
|
2YWV
 
 | | Crystal structure of SAICAR synthetase from Geobacillus kaustophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoribosylaminoimidazole succinocarboxamide synthetase, ... | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of SAICAR synthetase from Geobacillus kaustophilus
To be Published
|
|
2YWW
 
 | | Crystal structure of aspartate carbamoyltransferase regulatory chain from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of aspartate carbamoyltransferase regulatory chain from Methanocaldococcus jannaschii
To be Published
|
|
2YWB
 
 | | Crystal structure of GMP synthetase from Thermus thermophilus | | Descriptor: | GMP synthase [glutamine-hydrolyzing] | | Authors: | Baba, S, Kanagawa, M, Yanai, H, Ishii, T, Kuramitsu, S, Yokoyama, S, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-20 | | Release date: | 2007-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of GMP synthetase from Thermus thermophilus
To be Published
|
|
2YYT
 
 | | Crystal structure of uncharacterized conserved protein from Geobacillus kaustophilus | | Descriptor: | Orotidine 5'-phosphate decarboxylase | | Authors: | Kanagawa, M, Baba, S, Nakamura, Y, Bessho, Y, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-02 | | Release date: | 2007-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of uncharacterized conserved protein from Geobacillus kaustophilus
To be Published
|
|
2YZK
 
 | | Crystal structure of orotate phosphoribosyltransferase from Aeropyrum pernix | | Descriptor: | Orotate phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of orotate phosphoribosyltransferase from Aeropyrum pernix
To be Published
|
|
2Z01
 
 | | Crystal structure of phosphoribosylaminoimidazole synthetase from Geobacillus kaustophilus | | Descriptor: | Phosphoribosylformylglycinamidine cyclo-ligase | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and ligand binding of PurM proteins from Thermus thermophilus and Geobacillus kaustophilus
J.Biochem., 2015
|
|
2Z00
 
 | | Crystal structure of dihydroorotase from Thermus thermophilus | | Descriptor: | Dihydroorotase, ZINC ION | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of dihydroorotase from Thermus thermophilus
To be Published
|
|
